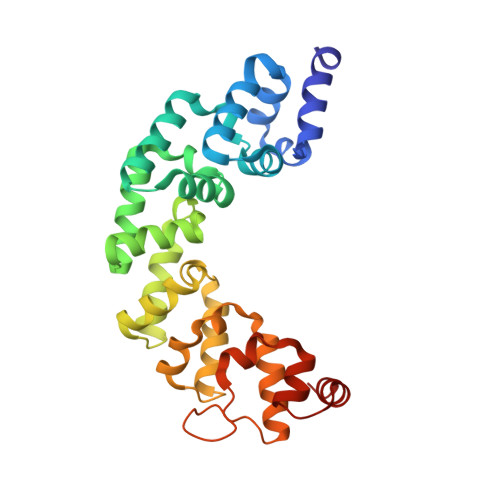
Structure of mitochondrial transcription termination factor 3 reveals a novel nucleic acid-binding domain.
Spahr, H., Samuelsson, T., Hallberg, B.M., Gustafsson, C.M.(2010) Biochem Biophys Res Commun 397: 386-390
- PubMed: 20430012
- DOI: https://doi.org/10.1016/j.bbrc.2010.04.130
- Primary Citation of Related Structures:
3M66 - PubMed Abstract:
In mammalian cells, a family of mitochondrial transcription termination factors (MTERFs) regulates mitochondrial gene expression. MTERF family members share a approximately 270 residues long MTERF-domain required for DNA binding and transcription regulation. However, the structure of this widely conserved domain is unknown. Here, we show that the MTERF-domain of human MTERF3 forms a half-doughnut-shaped right-handed superhelix. The superhelix is built from alpha-helical tandem repeats that display a novel triangular three-helix motif. This repeat motif, which we denote the MTERF-motif, is a conserved structural element present in proteins from metazoans, plants, and protozoans. Furthermore, a narrow, strongly positively charged nucleic acid-binding path is found in the middle of the concave side of the half-doughnut. This arrangement suggests a half clamp nucleic acid-binding mode for MTERF-domains.
- Dept. of Laboratory Medicine, Division of Metabolic Diseases, Karolinska Institutet, Stockholm, Sweden.
Organizational Affiliation: