Structural basis for the negative regulation of bacterial stress response by RseB
Kim, D.Y., Kwon, E., Choi, J.K., Hwang, H.-Y., Kim, K.K.(2010) Protein Sci 19: 1258-1263
- PubMed: 20512978
- DOI: https://doi.org/10.1002/pro.393
- Primary Citation of Related Structures:
3M4W - PubMed Abstract:
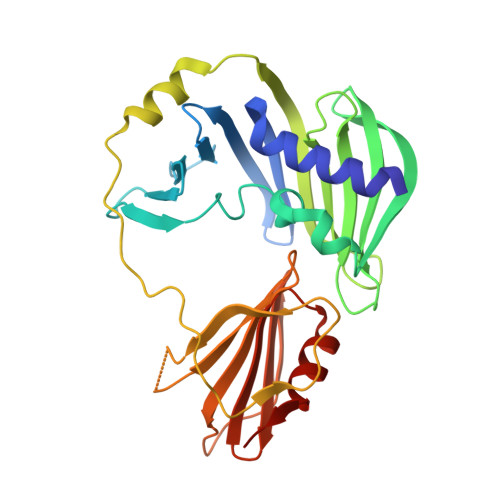
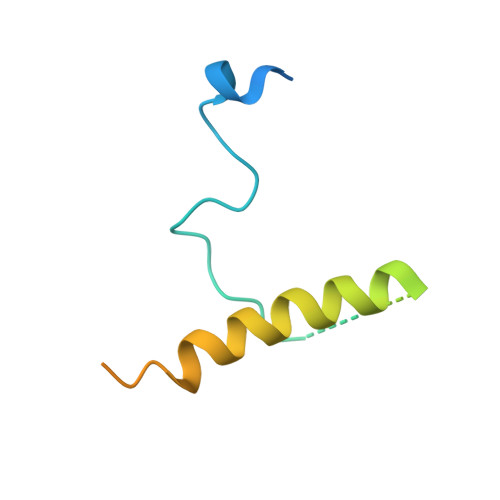
The sigmaE-dependent stress response in bacterial cells is initiated by the DegS- and RseP-regulated intramembrane proteolysis of a membrane-spanning antisigma factor, RseA. RseB binds to RseA and inhibits its sequential cleavage, thereby functioning as a negative modulator of this response. In the crystal structure of the periplasmic domain of RseA bound to RseB, the DegS cleavage site of RseA is unstructured, however, its P1 residue is buried in the hydrophobic pocket of RseB, which suggests that RseB binding blocks the access of DegS to the cleavage site.
- Department of Molecular Cell Biology, Sungkyunkwan University School of Medicine, Suwon 440-746, Korea.
Organizational Affiliation: