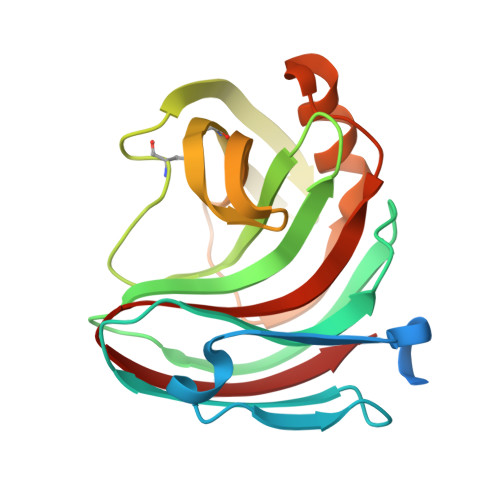
Structural insights into the acidophilic pH adaptation of a novel endo-1,4-beta-xylanase from Scytalidium acidophilum
Michaux, C., Pouyez, J., Mayard, A., Vandurm, P., Housen, I., Wouters, J.(2010) Biochimie 92: 1407-1415
- PubMed: 20621155
- DOI: https://doi.org/10.1016/j.biochi.2010.07.003
- Primary Citation of Related Structures:
3M4F - PubMed Abstract:
In this study, the crystal structure of a novel endo-1,4-β-xylanase from Scytalidium acidophilum, XYL1, was solved at 1.9Å resolution. This is one of the few solved crystal structures of acidophilic proteins. The enzyme has the overall fold typical to family 11 xylanases. Comparison of this structure with other homologous acidophilic, neutrophilic and alkalophilic xylanases provides additional insights into the general features involved in low pH adaptation (stability and activity). Several sequence and structure modifications appeared to be responsible for the acidophilic characteristic: (a) the presence of an aspartic acid H bonded to the acid/base catalyst (b) the nature of specifically conserved residues in the active site (c) the negative potential at the surface (d) the decreased number of salt bridges and H bonds in comparison with highly alkaline enzymes.
- Unité de Chimie Physique Structurale et Théorique, Chemistry Department, FUNDP, 61 rue de Bruxelles, B-5000 Namur, Belgium. catherine.michaux@fundp.ac.be
Organizational Affiliation: