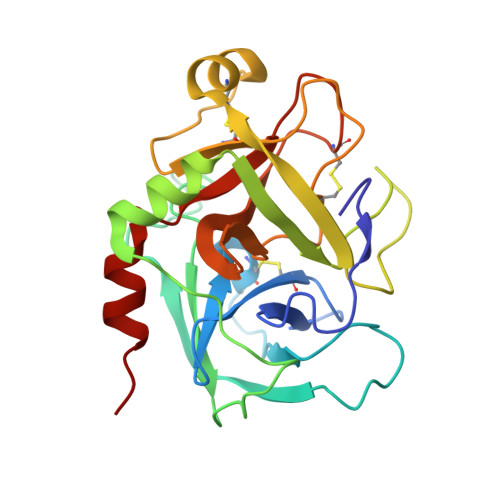
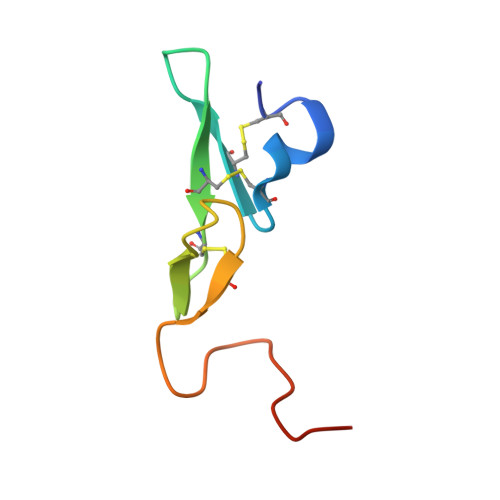
Structure Based Drug Design: Development of Potent and Selective Factor IXa (FIXa) Inhibitors.
Wang, S., Beck, R., Burd, A., Blench, T., Marlin, F., Ayele, T., Buxton, S., Dagostin, C., Malic, M., Joshi, R., Barry, J., Sajad, M., Cheung, C., Shaikh, S., Chahwala, S., Chander, C., Baumgartner, C., Holthoff, H.P., Murray, E., Blackney, M., Giddings, A.(2010) J Med Chem 53: 1473-1482
- PubMed: 20121197
- DOI: https://doi.org/10.1021/jm901476x
- Primary Citation of Related Structures:
3LC5 - PubMed Abstract:
On the basis of our understanding on the binding interactions of the benzothiophene template within the FIXa active site by X-ray crystallography and molecular modeling studies, we developed our SAR strategy by targeting the 4-position of the template to access the S1 beta and S2-S4 sites. A number of highly selective and potent factor Xa (FXa) and FIXa inhibitors were identified by simple switch of functional groups with conformational changes toward the S2-S4 sites.
- Department of Medicinal Chemistry, Trigen Ltd., Emmanuel Kaye Building, 1B Manresa Road,London SW3 6LR, UK. shouming_wang@yahoo.co.uk
Organizational Affiliation: