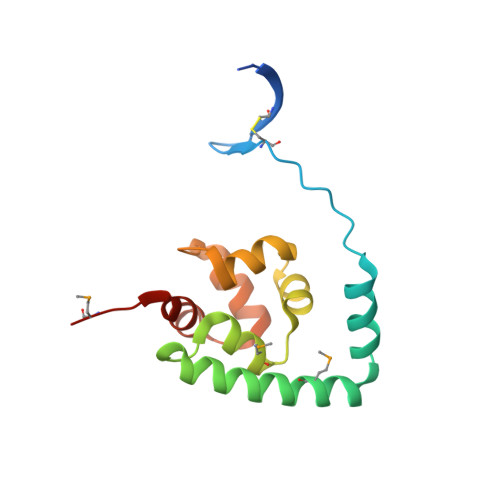
The crystal structure of NGO0477 from Neisseria gonorrhoeae reveals a novel protein fold incorporating a helix-turn-helix motif.
Ren, J., Sainsbury, S., Nettleship, J.E., Saunders, N.J., Owens, R.J.(2010) Proteins 78: 1798-1802
- PubMed: 20196080
- DOI: https://doi.org/10.1002/prot.22698
- Primary Citation of Related Structures:
3KXA - The Oxford Protein Production Facility and Division of Structural Biology, University of Oxford, Oxford OX3 7BN, United Kingdom.
Organizational Affiliation: