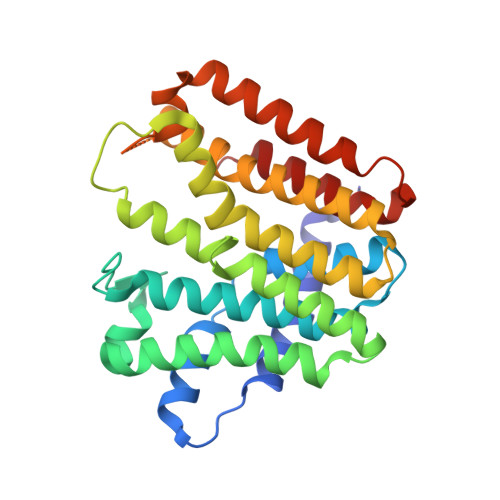
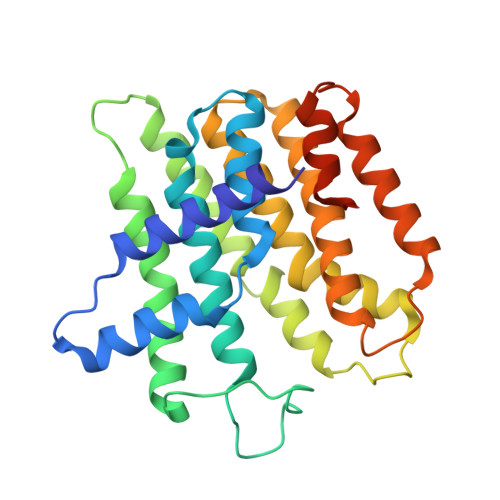
Structure of a heterotetrameric geranyl pyrophosphate synthase from mint (Mentha piperita) reveals intersubunit regulation
Chang, T.-H., Hsieh, F.-L., Ko, T.-P., Teng, K.-H., Liang, P.-H., Wang, A.H.-J.(2010) Plant Cell 22: 454-467
- PubMed: 20139160
- DOI: https://doi.org/10.1105/tpc.109.071738
- Primary Citation of Related Structures:
3KRA, 3KRC, 3KRF, 3KRO, 3KRP - PubMed Abstract:
Terpenes (isoprenoids), derived from isoprenyl pyrophosphates, are versatile natural compounds that act as metabolism mediators, plant volatiles, and ecological communicators. Divergent evolution of homomeric prenyltransferases (PTSs) has allowed PTSs to optimize their active-site pockets to achieve catalytic fidelity and diversity. Little is known about heteromeric PTSs, particularly the mechanisms regulating formation of specific products. Here, we report the crystal structure of the (LSU . SSU)(2)-type (LSU/SSU = large/small subunit) heterotetrameric geranyl pyrophosphate synthase (GPPS) from mint (Mentha piperita). The LSU and SSU of mint GPPS are responsible for catalysis and regulation, respectively, and this SSU lacks the essential catalytic amino acid residues found in LSU and other PTSs. Whereas no activity was detected for individually expressed LSU or SSU, the intact (LSU . SSU)(2) tetramer produced not only C(10)-GPP at the beginning of the reaction but also C(20)-GGPP (geranylgeranyl pyrophosphate) at longer reaction times. The activity for synthesizing C(10)-GPP and C(20)-GGPP, but not C(15)-farnesyl pyrophosphate, reflects a conserved active-site structure of the LSU and the closely related mustard (Sinapis alba) homodimeric GGPPS. Furthermore, using a genetic complementation system, we showed that no C(20)-GGPP is produced by the mint GPPS in vivo. Presumably through protein-protein interactions, the SSU remodels the active-site cavity of LSU for synthesizing C(10)-GPP, the precursor of volatile C(10)-monoterpenes.
- Institute of Biological Chemistry, Academia Sinica, Taipei 115, Taiwan. r93b46009@ntu.edu.tw
Organizational Affiliation: