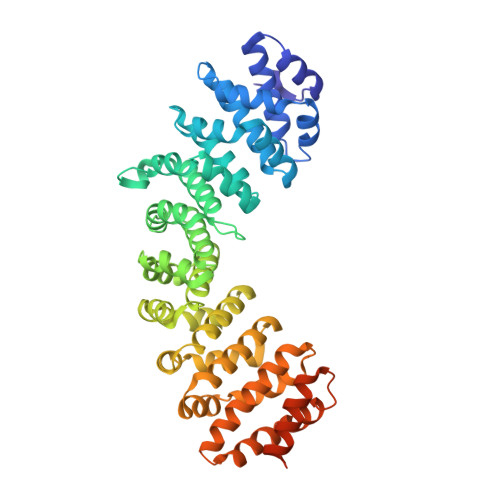
Novel binding of the mitotic regulator TPX2 (target protein for Xenopus kinesin-like protein 2) to importin-alpha.
Giesecke, A., Stewart, M.(2010) J Biological Chem 285: 17628-17635
- PubMed: 20335181
- DOI: https://doi.org/10.1074/jbc.M110.102343
- Primary Citation of Related Structures:
3KND - PubMed Abstract:
Several aspects of mitotic spindle assembly are orchestrated by the Ran GTPase through its modulation of the interaction between spindle assembly factors and importin-alpha. One such factor is TPX2 that promotes microtubule assembly in the vicinity of chromosomes. TPX2 is inhibited when bound to importin-alpha, which occurs when the latter is bound to importin-beta. The importin-alpha:beta interaction is disrupted by the high RanGTP concentration near the chromosomes, releasing TPX2. In more distal regions, where Ran is predominantly GDP-bound, TPX2 remains bound to importin-alpha and so is inhibited. Here we use a combination of structural and biochemical methods to define the basis for TPX2 binding to importin-alpha. A 2.2 A resolution crystal structure shows that the primary nuclear localization signal ((284)KRKH(287)) of TPX2, which has been shown to be crucial for inhibition, binds to the minor NLS-binding site on importin-alpha. This atypical interaction pattern was confirmed using complementary binding studies that employed importin-alpha variants in which binding to either the major or minor NLS-binding site was impaired, together with competition assays using the SV40 monopartite NLS that binds primarily to the major site. The different way in which TPX2 binds to importin-alpha could account for much of the selectivity necessary during mitosis because this would reduce the competition for binding to importin-alpha from other NLS-containing proteins.
- Medical Research Council Laboratory of Molecular Biology, Cambridge CB2 0QH, United Kingdom.
Organizational Affiliation: