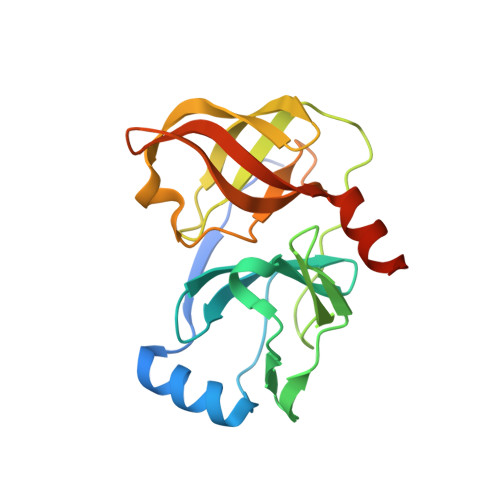
P4 capped amides and lactams as HCV NS3 protease inhibitors with improved potency and DMPK profile.
Nair, L.G., Sannigrahi, M., Bogen, S., Pinto, P., Chen, K.X., Prongay, A., Tong, X., Cheng, K.C., Girijavallabhan, V., George Njoroge, F.(2010) Bioorg Med Chem Lett 20: 567-570
- PubMed: 20004570
- DOI: https://doi.org/10.1016/j.bmcl.2009.11.094
- Primary Citation of Related Structures:
3KN2 - PubMed Abstract:
SAR studies on the extension of P3 unit of Boceprevir (1, SCH 503034) with amides and lactams and their synthesis is described. Extensive SAR studies resulted in the identification of 36 bearing 4, 4-dimethyl lactam as the new P4 cap unit with improved potency (K(i)( *)=15nM, EC 90=70nM) and pharmacokinetic properties (Rat AUC (PO)=3.52microMh) compared to 1.
- Chemical Research, Merck Research Laboratories, 2015 Galloping Hill Road, Kenilworth, NJ 07033, USA. latha.nair@spcorp.com
Organizational Affiliation: