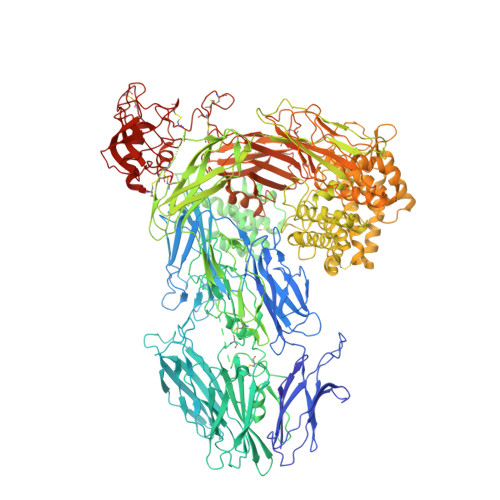
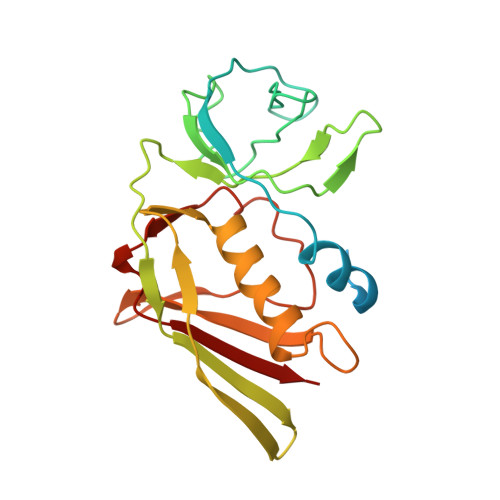
Structural basis for inhibition of complement C5 by the SSL7 protein from Staphylococcus aureus
Laursen, N.S., Gordon, N., Hermans, S., Lorenz, N., Jackson, N., Wines, B., Spillner, E., Christensen, J.B., Jensen, M., Fredslund, F., Bjerre, M., Sottrup-Jensen, L., Fraser, J.D., Andersen, G.R.(2010) Proc Natl Acad Sci U S A 107: 3681-3686
- PubMed: 20133685
- DOI: https://doi.org/10.1073/pnas.0910565107
- Primary Citation of Related Structures:
3KLS, 3KM9 - PubMed Abstract:
Staphylococcus aureus secretes the SSL7 protein as part of its immune evasion strategy. The protein binds both complement C5 and IgA, yet it is unclear whether SSL7 cross-links these two proteins and, if so, what purpose this serves the pathogen. We have isolated a stable IgA-SSL7-C5 complex, and our crystal structure of the C5-SSL7 complex confirms that binding to C5 occurs exclusively through the C-terminal beta-grasp domain of SSL7 leaving the OB domain free to interact with IgA. SSL7 interacts with C5 >70 A from the C5a cleavage site without inducing significant conformational changes in C5, and efficient inhibition of convertase cleavage of C5 is shown to be IgA dependent. Inhibition of C5a production and bacteriolysis are all shown to require C5 and IgA binding while inhibition of hemolysis is achieved by the C5 binding SSL7 beta-grasp domain alone. These results provide a conceptual and structural basis for the development of a highly specific complement inhibitor preventing only the formation of the lytic membrane attack complex without affecting the important signaling functions of C5a.
- Department of Molecular Biology, University of Aarhus, DK-8000 Aarhus, Denmark.
Organizational Affiliation: