Crystal structure of functionally unknown HSP33 from Saccharomyces cerevisiae
Hwang, K.Y., Sung, M.W., Lee, W.H.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Probable chaperone protein HSP33 | 244 | Saccharomyces cerevisiae | Mutation(s): 0 Gene Names: HSP33, YOR391C EC: 3.2 (PDB Primary Data), 4.2.1.130 (UniProt) | 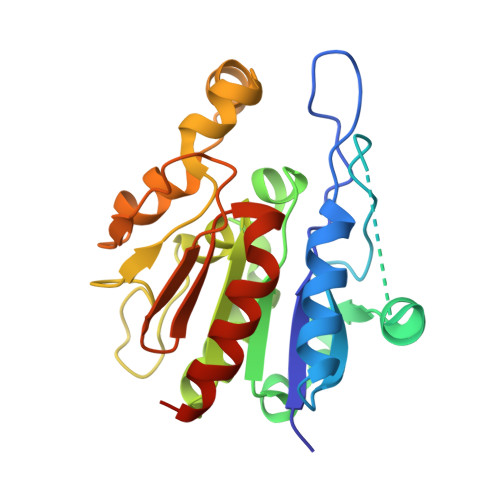 | |
UniProt | |||||
Find proteins for Q08914 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore Q08914 Go to UniProtKB: Q08914 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q08914 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 43.45 | α = 90 |
| b = 68.738 | β = 100.08 |
| c = 66.315 | γ = 90 |
| Software Name | Purpose |
|---|---|
| ADSC | data collection |
| CNS | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| CNS | phasing |