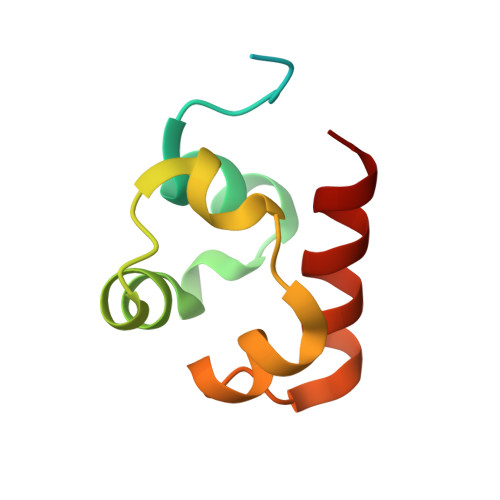
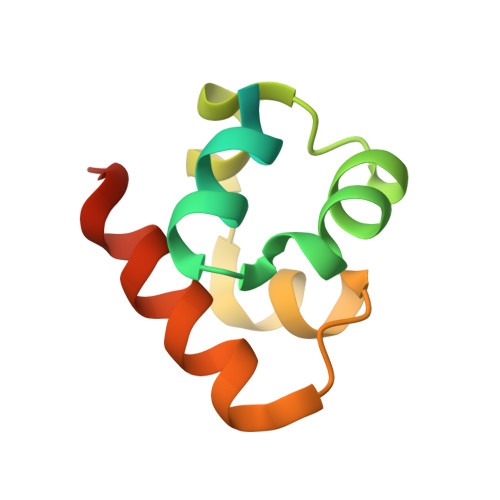
Co-Crystal Structure of the SAM Domains of Human Ephrin Type-A Receptor 1 and Human Ephrin Type-A Receptor 2
Walker, J.R., Yermekbayeva, L., Butler-Cole, C., Weigelt, J., Bountra, C., Arrowsmith, C.H., Edwards, A.M., Bochkarev, A., Dhe-Paganon, S.To be published.