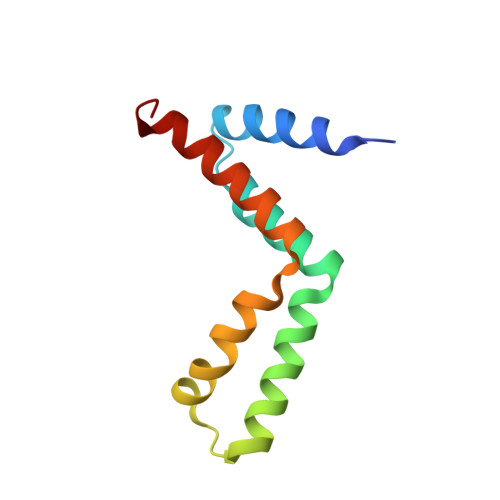
Structural basis for the interaction between yeast Spt-Ada-Gcn5 acetyltransferase (SAGA) complex components Sgf11 and Sus1
Ellisdon, A.M., Jani, D., Kohler, A., Hurt, E., Stewart, M.(2010) J Biological Chem 285: 3850-3856
- PubMed: 20007317
- DOI: https://doi.org/10.1074/jbc.M109.070839
- Primary Citation of Related Structures:
3KIK, 3KJL - PubMed Abstract:
Sus1 is a central component of the yeast gene gating machinery, the process by which actively transcribing genes such as GAL1 become associated with nuclear pore complexes. Sus1 is a component of both the SAGA transcriptional co-activator complex and the TREX-2 complex that binds to nuclear pore complexes. TREX-2 contains two Sus1 chains that have an articulated helical hairpin fold, enabling them to wrap around an extended alpha-helix in Sac3, following a helical hydrophobic stripe. In SAGA, Sus1 binds to Sgf11 and has been proposed to provide a link between SAGA and TREX-2. We present here the crystal structure of the complex between Sus1 and the N-terminal region of Sgf11 that forms an extended alpha-helix around which Sus1 wraps in a manner that shares some similarities with the Sus1-Sac3 interface in TREX-2. However, the Sus1-binding site on Sgf11 is somewhat shorter than on Sac3 and is based on a narrower hydrophobic stripe. Engineered mutants that disrupt the Sgf11-Sus1 interaction in vitro confirm the importance of the hydrophobic helical stripe in molecular recognition. Helix alpha1 of the Sus1-articulated hairpin does not bind directly to Sgf11 and adopts a wide range of conformations within and between crystal forms, consistent with the presence of a flexible hinge and also with results from previous extensive mutagenesis studies (Klöckner, C., Schneider, M., Lutz, S., Jani, D., Kressler, D., Stewart, M., Hurt, E., and Köhler, A. (2009) J. Biol. Chem. 284, 12049-12056). A single Sus1 molecule cannot bind Sgf11 and Sac3 simultaneously and this, combined with the structure of the Sus1-Sgf11 complex, indicates that Sus1 forms separate subcomplexes within SAGA and TREX-2.
- From the Medical Research Council Laboratory of Molecular Biology, Hills Road, Cambridge CB2 0QH, United Kingdom and.
Organizational Affiliation: