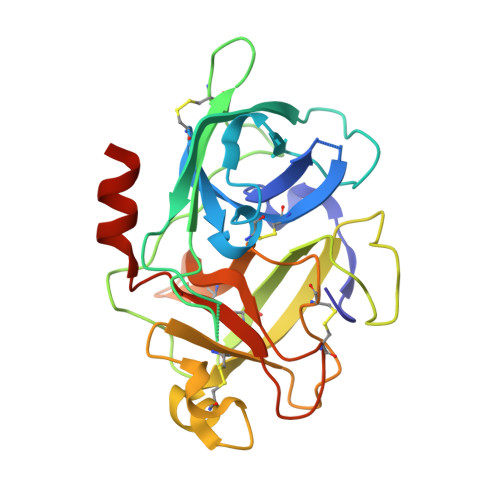
Crystal Structures of 2-Aminobenzothiazole-based Inhibitors in Complexes with Urokinase-type Plasminogen Activator
Jiang, L.-G., Yu, H.-Y., Yuan, C., Wang, J.-D., Chen, L.-Q., Meehan, E.J., Huang, Z.-X., Huang, M.-D.(2009) Chin J Struct Chem 28: 1427-1432