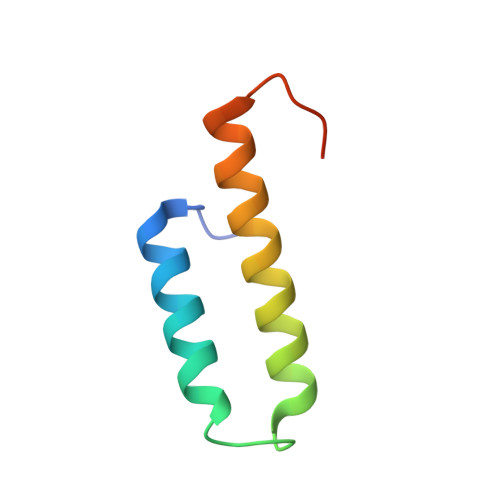
Structure of the GLD-1 homodimerization domain: insights into STAR protein-mediated translational regulation.
Beuck, C., Szymczyna, B.R., Kerkow, D.E., Carmel, A.B., Columbus, L., Stanfield, R.L., Williamson, J.R.(2010) Structure 18: 377-389
- PubMed: 20223220
- DOI: https://doi.org/10.1016/j.str.2009.12.016
- Primary Citation of Related Structures:
3K6T, 3KBL - PubMed Abstract:
Posttranscriptional regulation of gene expression is an important mechanism for modulating protein levels in eukaryotes, especially in developmental pathways. The highly conserved homodimeric STAR/GSG proteins play a key role in regulating translation by binding bipartite consensus sequences in the untranslated regions of target mRNAs, but the exact mechanism remains unknown. Structures of STAR protein RNA binding subdomains have been determined, but structural information is lacking for the homodimerization subdomain. Here, we present the structure of the C. elegans GLD-1 homodimerization domain dimer, determined by a combination of X-ray crystallography and NMR spectroscopy, revealing a helix-turn-helix monomeric fold with the two protomers stacked perpendicularly. Structure-based mutagenesis demonstrates that the dimer interface is not easily disrupted, but the structural integrity of the monomer is crucial for GLD-1 dimerization. Finally, an improved model for STAR-mediated translational regulation of mRNA, based on the GLD-1 homodimerization domain structure, is presented.
- Department of Molecular Biology, Department of Chemistry, and Skaggs Institute for Chemical Biology, The Scripps Research Institute, La Jolla, CA 92037, USA.
Organizational Affiliation: