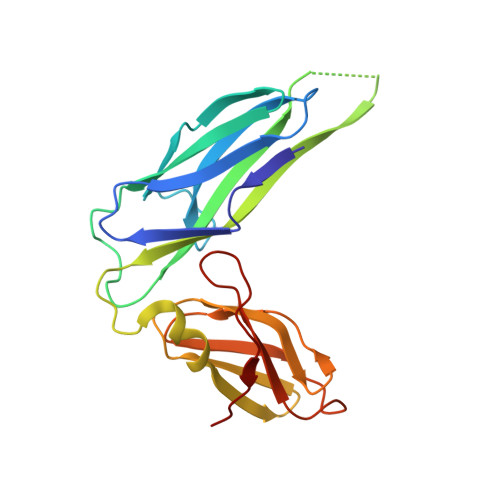
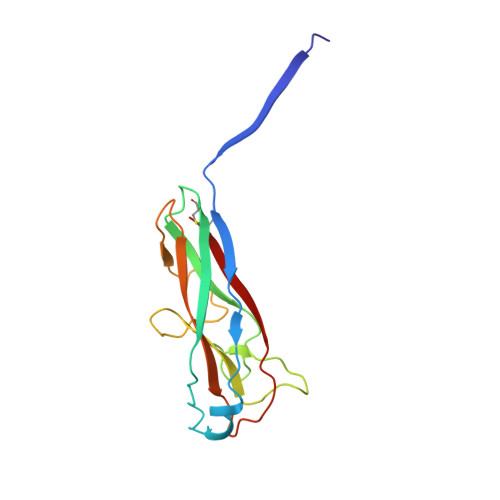
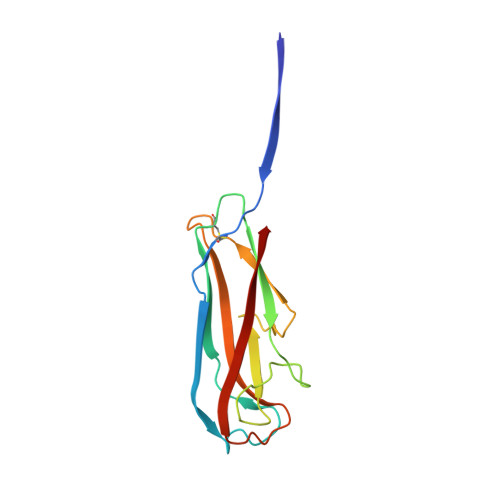
Structural basis for mechanical force regulation of the adhesin FimH via finger trap-like beta sheet twisting.
Le Trong, I., Aprikian, P., Kidd, B.A., Forero-Shelton, M., Tchesnokova, V., Rajagopal, P., Rodriguez, V., Interlandi, G., Klevit, R., Vogel, V., Stenkamp, R.E., Sokurenko, E.V., Thomas, W.E.(2010) Cell 141: 645-655
- PubMed: 20478255
- DOI: https://doi.org/10.1016/j.cell.2010.03.038
- Primary Citation of Related Structures:
3JWN - PubMed Abstract:
The Escherichia coli fimbrial adhesive protein, FimH, mediates shear-dependent binding to mannosylated surfaces via force-enhanced allosteric catch bonds, but the underlying structural mechanism was previously unknown. Here we present the crystal structure of FimH incorporated into the multiprotein fimbrial tip, where the anchoring (pilin) domain of FimH interacts with the mannose-binding (lectin) domain and causes a twist in the beta sandwich fold of the latter. This loosens the mannose-binding pocket on the opposite end of the lectin domain, resulting in an inactive low-affinity state of the adhesin. The autoinhibition effect of the pilin domain is removed by application of tensile force across the bond, which separates the domains and causes the lectin domain to untwist and clamp tightly around the ligand like a finger-trap toy. Thus, beta sandwich domains, which are common in multidomain proteins exposed to tensile force in vivo, can undergo drastic allosteric changes and be subjected to mechanical regulation.
- Department of Biological Structure, Box 357420, University of Washington, Seattle, WA 98195, USA.
Organizational Affiliation: