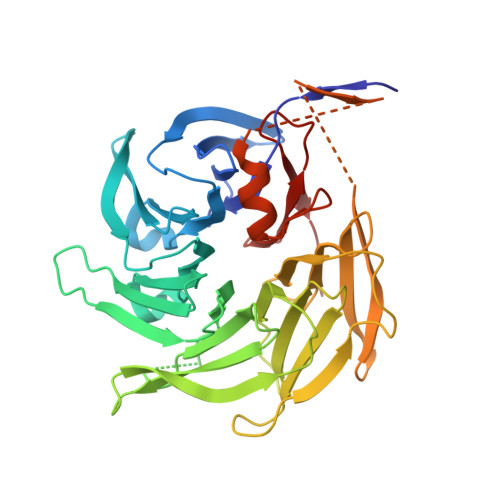
Molecular architecture of the Nup84-Nup145C-Sec13 edge element in the nuclear pore complex lattice.
Brohawn, S.G., Schwartz, T.U.(2009) Nat Struct Mol Biol 16: 1173-1177
- PubMed: 19855394
- DOI: https://doi.org/10.1038/nsmb.1713
- Primary Citation of Related Structures:
3JRO, 3JRP - PubMed Abstract:
Nuclear pore complexes (NPCs) facilitate all nucleocytoplasmic transport. These massive protein assemblies are modular, with a stable structural scaffold supporting more dynamically attached components. The scaffold is made from multiple copies of the heptameric Y complex and the heteromeric Nic96 complex. We previously showed that members of these core subcomplexes specifically share an ACE1 fold with Sec31 of the COPII vesicle coat, and we proposed a lattice model for the NPC based on this commonality. Here we present the crystal structure of the heterotrimeric 134-kDa complex of Nup84-Nup145C-Sec13 of the Y complex. The heterotypic ACE1 interaction of Nup84 and Nup145C is analogous to the homotypic ACE1 interaction of Sec31 that forms COPII lattice edge elements and is inconsistent with the alternative 'fence-like' NPC model. We construct a molecular model of the Y complex and compare the architectural principles of COPII and NPC lattices.
- Department of Biology, Massachusetts Institute of Technology, Cambridge, Massachusetts, USA.
Organizational Affiliation: