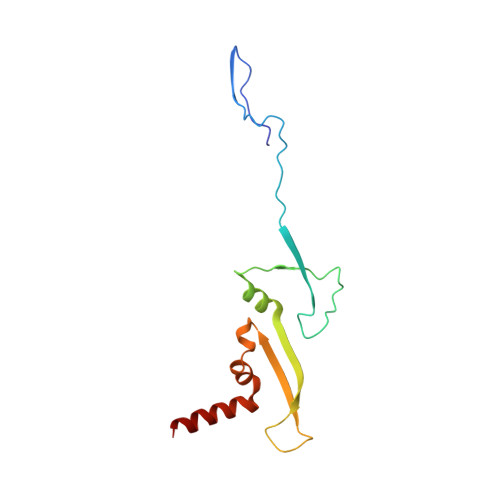
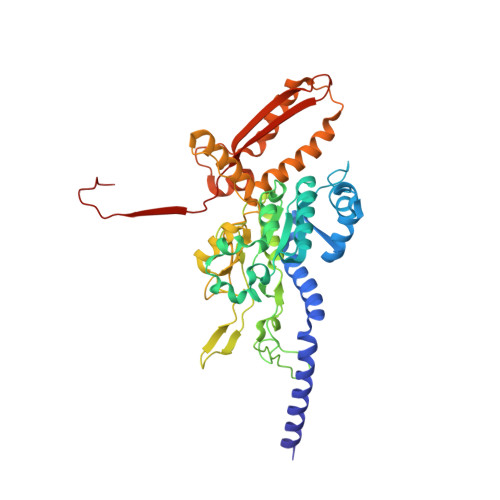
Structure of the Type VI Secretion System Contractile Sheath.
Kudryashev, M., Wang, R.Y., Brackmann, M., Scherer, S., Maier, T., Baker, D., DiMaio, F., Stahlberg, H., Egelman, E.H., Basler, M.(2015) Cell 160: 952-962
- PubMed: 25723169
- DOI: https://doi.org/10.1016/j.cell.2015.01.037
- Primary Citation of Related Structures:
3J9G - PubMed Abstract:
Bacteria use rapid contraction of a long sheath of the type VI secretion system (T6SS) to deliver effectors into a target cell. Here, we present an atomic-resolution structure of a native contracted Vibrio cholerae sheath determined by cryo-electron microscopy. The sheath subunits, composed of tightly interacting proteins VipA and VipB, assemble into a six-start helix. The helix is stabilized by a core domain assembled from four β strands donated by one VipA and two VipB molecules. The fold of inner and middle layers is conserved between T6SS and phage sheaths. However, the structure of the outer layer is distinct and suggests a mechanism of interaction of the bacterial sheath with an accessory ATPase, ClpV, that facilitates multiple rounds of effector delivery. Our results provide a mechanistic insight into assembly of contractile nanomachines that bacteria and phages use to translocate macromolecules across membranes.
- Focal Area Infection Biology, Biozentrum, University of Basel, Klingelbergstrasse 50/70, CH-4056 Basel, Switzerland; Center for Cellular Imaging and NanoAnalytics, Biozentrum, University of Basel, Mattenstrasse 26, CH-4058 Basel, Switzerland.
Organizational Affiliation: