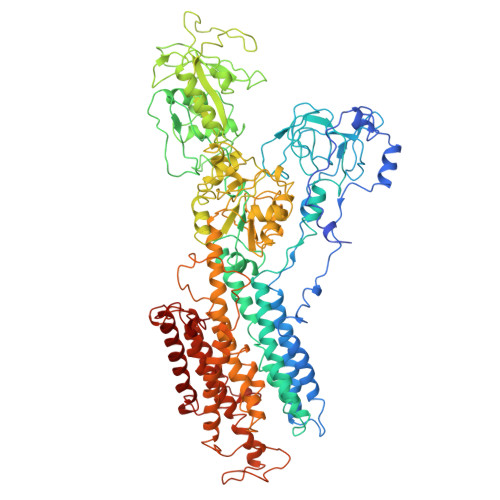
Inter-subunit interaction of gastric H+,K+-ATPase prevents reverse reaction of the transport cycle
Abe, K., Tani, K., Nishizawa, T., Fujiyoshi, Y.(2009) EMBO J 28: 1637-1643
- PubMed: 19387495
- DOI: https://doi.org/10.1038/emboj.2009.102
- Primary Citation of Related Structures:
3IXZ - PubMed Abstract:
The gastric H(+),K(+)-ATPase is an ATP-driven proton pump responsible for generating a million-fold proton gradient across the gastric membrane. We present the structure of gastric H(+),K(+)-ATPase at 6.5 A resolution as determined by electron crystallography of two-dimensional crystals. The structure shows the catalytic alpha-subunit and the non-catalytic beta-subunit in a pseudo-E(2)P conformation. Different from Na(+),K(+)-ATPase, the N-terminal tail of the beta-subunit is in direct contact with the phosphorylation domain of the alpha-subunit. This interaction may hold the phosphorylation domain in place, thus stabilizing the enzyme conformation and preventing the reverse reaction of the transport cycle. Indeed, truncation of the beta-subunit N-terminus allowed the reverse reaction to occur. These results suggest that the beta-subunit N-terminus prevents the reverse reaction from E(2)P to E(1)P, which is likely to be relevant for the generation of a large H(+) gradient in vivo situation.
- Department of Biophysics, Faculty of Science, Kyoto University, Kyoto, Japan.
Organizational Affiliation: