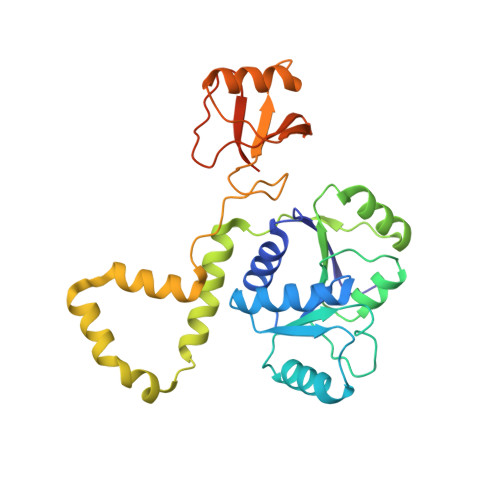
The Thermus thermophilus DEAD box helicase Hera contains a modified RNA recognition motif domain loosely connected to the helicase core.
Rudolph, M.G., Klostermeier, D.(2009) RNA 15: 1993-2001
- PubMed: 19710183
- DOI: https://doi.org/10.1261/rna.1820009
- Primary Citation of Related Structures:
3I31, 3I32 - PubMed Abstract:
DEAD box family helicases consist of a helicase core that is formed by two flexibly linked RecA-like domains. The helicase activity can be regulated by N- or C-terminal extensions flanking the core. Thermus thermophilus heat resistant RNA-dependent ATPase (Hera) is the first DEAD box helicase that forms a dimer using a unique dimerization domain. In addition to the dimerization domain, Hera contains a C-terminal RNA binding domain (RBD) that shares sequence homology only to uncharacterized proteins of the Deinococcus/Thermus group. The crystal structure of Hera_RBD reveals the fold of an altered RNA recognition motif (RRM) with limited structural homology to the RBD of the DEAD box helicase YxiN from Bacillus subtilis. Comparison with RRM/RNA complexes shows that a RNA binding mode different than that suggested for YxiN, but similar to U1A, can be inferred for Hera. The orientation of the RBD relative to the helicase core was defined in a second crystal structure of a Hera fragment including the C-terminal RecA domain, the dimerization domain, and the RBD. The structures allow construction of a model for the entire Hera helicase dimer. A likely binding surface for large RNA substrates that spans both RecA-like domains and the RBD is identified.
- Department of Molecular Structural Biology, University of Göttingen, D-37077 Göttingen, Germany.
Organizational Affiliation: