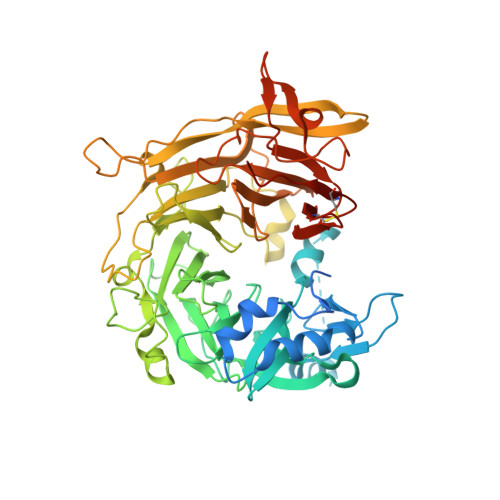
Crystal structure analysis reveals Pseudomonas PilY1 as an essential calcium-dependent regulator of bacterial surface motility.
Orans, J., Johnson, M.D., Coggan, K.A., Sperlazza, J.R., Heiniger, R.W., Wolfgang, M.C., Redinbo, M.R.(2010) Proc Natl Acad Sci U S A 107: 1065-1070
- PubMed: 20080557
- DOI: https://doi.org/10.1073/pnas.0911616107
- Primary Citation of Related Structures:
3HX6 - PubMed Abstract:
Several bacterial pathogens require the "twitching" motility produced by filamentous type IV pili (T4P) to establish and maintain human infections. Two cytoplasmic ATPases function as an oscillatory motor that powers twitching motility via cycles of pilus extension and retraction. The regulation of this motor, however, has remained a mystery. We present the 2.1 A resolution crystal structure of the Pseudomonas aeruginosa pilus-biogenesis factor PilY1, and identify a single site on this protein required for bacterial translocation. The structure reveals a modified beta-propeller fold and a distinct EF-hand-like calcium-binding site conserved in pathogens with retractile T4P. We show that preventing calcium binding by PilY1 using either an exogenous calcium chelator or mutation of a single residue disrupts Pseudomonas twitching motility by eliminating surface pili. In contrast, placing a lysine in this site to mimic the charge of a bound calcium interferes with motility in the opposite manner--by producing an abundance of nonfunctional surface pili. Our data indicate that calcium binding and release by the unique loop identified in the PilY1 crystal structure controls the opposing forces of pilus extension and retraction. Thus, PilY1 is an essential, calcium-dependent regulator of bacterial twitching motility.
- Department of Chemistry, University of North Carolina at Chapel Hill, CB 3290, Chapel Hill, NC 27599, USA.
Organizational Affiliation: