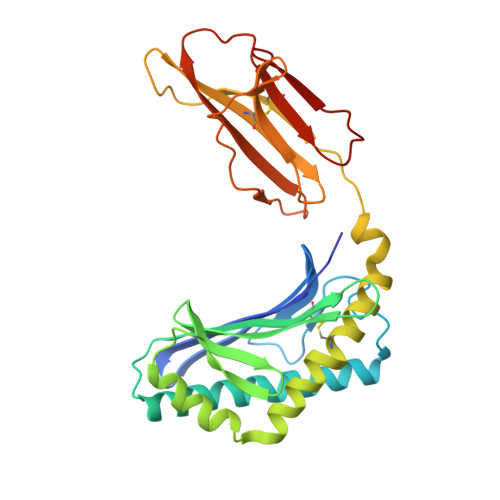
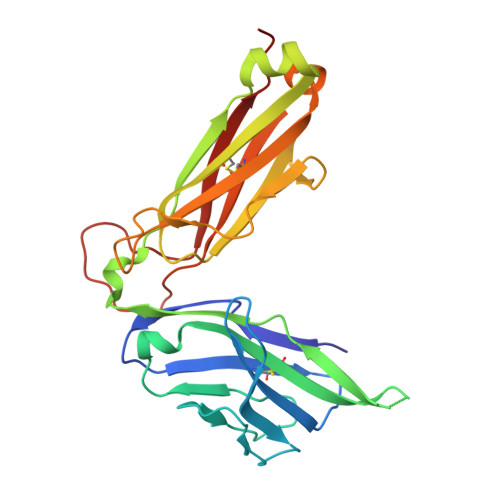
Differential recognition of CD1d-alpha-galactosyl ceramide by the V beta 8.2 and V beta 7 semi-invariant NKT T cell receptors
Pellicci, D.G., Patel, O., Kjer-Nielsen, L., Pang, S.S., Sullivan, L.C., Kyparissoudis, K., Brooks, A.G., Reid, H.H., Gras, S., Lucet, I.S., Koh, R., Smyth, M.J., Mallevaey, T., Matsuda, J.L., Gapin, L., McCluskey, J., Godfrey, D.I., Rossjohn, J.(2009) Immunity 31: 47-59
- PubMed: 19592275
- DOI: https://doi.org/10.1016/j.immuni.2009.04.018
- Primary Citation of Related Structures:
3HE6, 3HE7, 3HUJ - PubMed Abstract:
The semi-invariant natural killer T cell receptor (NKT TCR) recognizes CD1d-lipid antigens. Although the TCR alpha chain is typically invariant, the beta chain expression is more diverse, where three V beta chains are commonly expressed in mice. We report the structures of V alpha 14-V beta 8.2 and V alpha 14-V beta 7 NKT TCRs in complex with CD1d-alpha-galactosylceramide (alpha-GalCer) and the 2.5 A structure of the human NKT TCR-CD1d-alpha-GalCer complex. Both V beta 8.2 and V beta 7 NKT TCRs and the human NKT TCR ligated CD1d-alpha-GalCer in a similar manner, highlighting the evolutionarily conserved interaction. However, differences within the V beta domains of the V beta 8.2 and V beta 7 NKT TCR-CD1d complexes resulted in altered TCR beta-CD1d-mediated contacts and modulated recognition mediated by the invariant alpha chain. Mutagenesis studies revealed the differing contributions of V beta 8.2 and V beta 7 residues within the CDR2 beta loop in mediating contacts with CD1d. Collectively we provide a structural basis for the differential NKT TCR V beta usage in NKT cells.
- Department of Microbiology and Immunology, University of Melbourne, Parkville, Victoria 3010, Australia.
Organizational Affiliation: