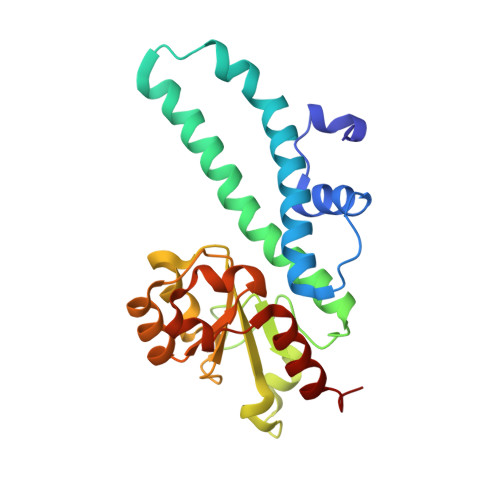
Structural mechanisms for the 5'-CCWGG sequence recognition by the N- and C-terminal domains of EcoRII.
Golovenko, D., Manakova, E., Tamulaitiene, G., Grazulis, S., Siksnys, V.(2009) Nucleic Acids Res 37: 6613-6624
- PubMed: 19729506
- DOI: https://doi.org/10.1093/nar/gkp699
- Primary Citation of Related Structures:
3HQF, 3HQG - PubMed Abstract:
EcoRII restriction endonuclease is specific for the 5'-CCWGG sequence (W stands for A or T); however, it shows no activity on a single recognition site. To activate cleavage it requires binding of an additional target site as an allosteric effector. EcoRII dimer consists of three structural units: a central catalytic core, made from two copies of the C-terminal domain (EcoRII-C), and two N-terminal effector DNA binding domains (EcoRII-N). Here, we report DNA-bound EcoRII-N and EcoRII-C structures, which show that EcoRII combines two radically different structural mechanisms to interact with the effector and substrate DNA. The catalytic EcoRII-C dimer flips out the central T:A base pair and makes symmetric interactions with the CC:GG half-sites. The EcoRII-N effector domain monomer binds to the target site asymmetrically in a single defined orientation which is determined by specific hydrogen bonding and van der Waals interactions with the central T:A pair in the major groove. The EcoRII-N mode of the target site recognition is shared by the large class of higher plant transcription factors of the B3 superfamily.
- Institute of Biotechnology, Graiciuno 8, LT-02241 Vilnius, Lithuania.
Organizational Affiliation: