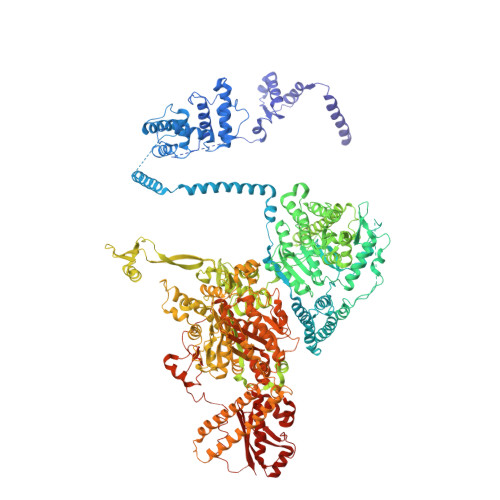
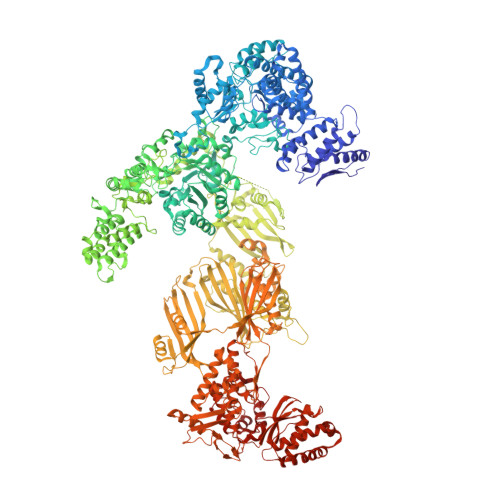
Multimeric options for the auto-activation of the Saccharomyces cerevisiae FAS type I megasynthase
Johansson, P., Mulinacci, B., Koestler, C., Vollrath, R., Oesterhelt, D., Grininger, M.(2009) Structure 17: 1063-1074
- PubMed: 19679086
- DOI: https://doi.org/10.1016/j.str.2009.06.014
- Primary Citation of Related Structures:
2WAS, 2WAT, 3HMJ - PubMed Abstract:
The fungal type I fatty acid synthase (FAS) is a 2.6 MDa multienzyme complex, catalyzing all necessary steps for the synthesis of long acyl chains. To be catalytically competent, the FAS must be activated by a posttranslational modification of the central acyl carrier domain (ACP) by an intrinsic phosphopantetheine transferase (PPT). However, recent X-ray structures of the fungal FAS revealed a barrel-shaped architecture, with PPT located at the outside of the barrel wall, spatially separated from the ACP caged in the inner volume. This separation indicated that the activation has to proceed before the assembly to the mature complex, in a conformation where the ACP and PPT domains can meet. To gain insight into the auto-activation reaction and also into the fungal FAS assembly pathway, we structurally and functionally characterized the Saccharomyces cerevisiae FAS type I PPT as part of the multienzyme protein and as an isolated domain.
- Department of Membrane Biochemistry, Max-Planck-Institute of Biochemistry, Martinsried, Germany. johansso@biochem.mpg.de
Organizational Affiliation: