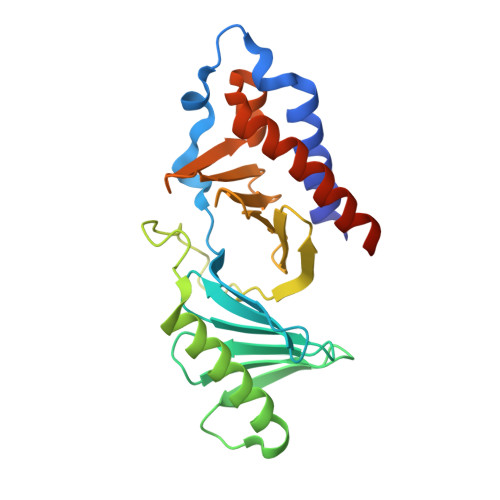
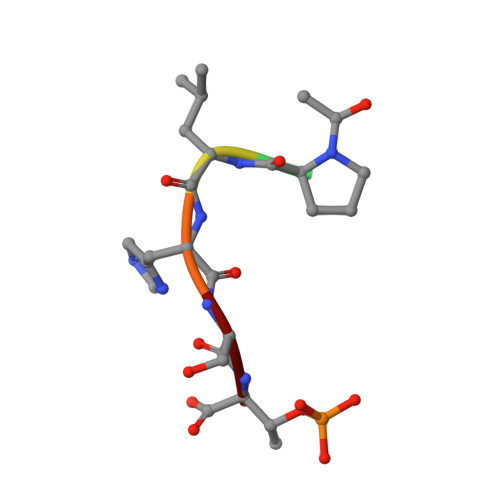
Structural and functional analyses of minimal phosphopeptides targeting the polo-box domain of polo-like kinase 1.
Yun, S.M., Moulaei, T., Lim, D., Bang, J.K., Park, J.E., Shenoy, S.R., Liu, F., Kang, Y.H., Liao, C., Soung, N.K., Lee, S., Yoon, D.Y., Lim, Y., Lee, D.H., Otaka, A., Appella, E., McMahon, J.B., Nicklaus, M.C., Burke, T.R., Yaffe, M.B., Wlodawer, A., Lee, K.S.(2009) Nat Struct Mol Biol 16: 876-882
- PubMed: 19597481
- DOI: https://doi.org/10.1038/nsmb.1628
- Primary Citation of Related Structures:
3C5L, 3FVH, 3HIH, 3HIK - PubMed Abstract:
Polo-like kinase-1 (Plk1) has a pivotal role in cell proliferation and is considered a potential target for anticancer therapy. The noncatalytic polo-box domain (PBD) of Plk1 forms a phosphoepitope binding module for protein-protein interaction. Here, we report the identification of minimal phosphopeptides that specifically interact with the PBD of human PLK1, but not those of the closely related PLK2 and PLK3. Comparative binding studies and analyses of crystal structures of the PLK1 PBD in complex with the minimal phosphopeptides revealed that the C-terminal SpT dipeptide functions as a high-affinity anchor, whereas the N-terminal residues are crucial for providing specificity and affinity to the interaction. Inhibition of the PLK1 PBD by phosphothreonine mimetic peptides was sufficient to induce mitotic arrest and apoptotic cell death. The mode of interaction between the minimal peptide and PBD may provide a template for designing therapeutic agents that target PLK1.
- Laboratory of Metabolism, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, Maryland, USA.
Organizational Affiliation: