The high-resolution structure of a Cobalt containing nitrile hydratase
Van Wyk, J.C., Tastan Bishop, A.O., Cowan, D.A., Sayed, M.F., Tsekoa, T.L., Sewell, B.T.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Nitrile hydratase alpha subunit | 216 | Aeribacillus pallidus | Mutation(s): 1 EC: 4.2.1.84 | 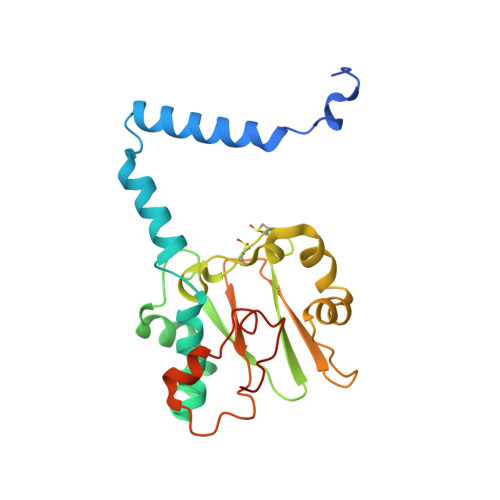 | |
UniProt | |||||
Find proteins for Q84FS5 (Bacillus sp. RAPc8) Explore Q84FS5 Go to UniProtKB: Q84FS5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q84FS5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Nitrile hydratase beta subunit | 229 | Aeribacillus pallidus | Mutation(s): 3 EC: 4.2.1.84 | 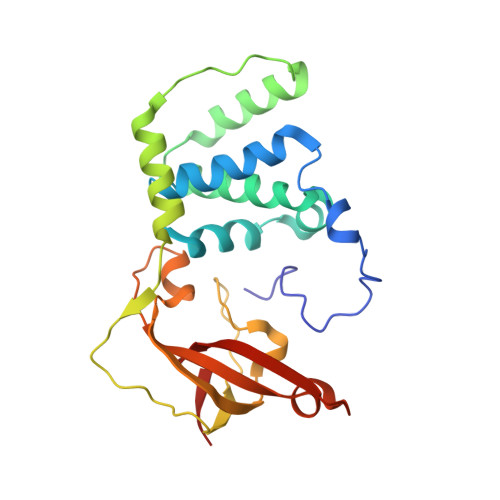 | |
UniProt | |||||
Find proteins for Q84FS6 (Bacillus sp. RAPc8) Explore Q84FS6 Go to UniProtKB: Q84FS6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q84FS6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 3CO Query on 3CO | C [auth A] | COBALT (III) ION Co JAWGVVJVYSANRY-UHFFFAOYSA-N |  | ||
| CL Query on CL | D [auth A], E [auth B] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| CSD Query on CSD | A | L-PEPTIDE LINKING | C3 H7 N O4 S |  | CYS |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 105.832 | α = 90 |
| b = 105.832 | β = 90 |
| c = 83.723 | γ = 90 |
| Software Name | Purpose |
|---|---|
| d*TREK | data scaling |
| PHASER | phasing |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| MxCuBE | data collection |
| d*TREK | data reduction |