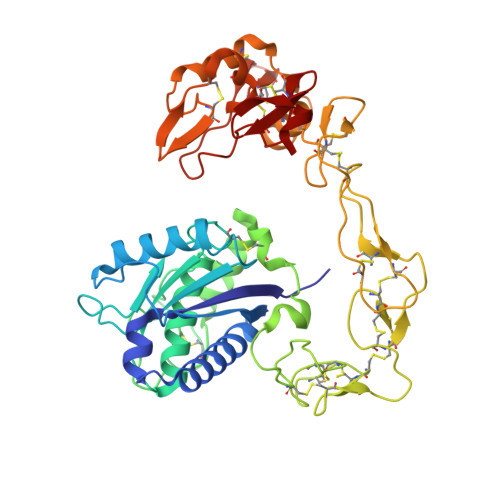
Structural basis of the autolysis of AaHIV suggests a novel target recognizing model for ADAM/reprolysin family proteins
Zhu, Z., Gao, Y., Zhu, Z., Yu, Y., Zhang, X., Zang, J., Teng, M., Niu, L.(2009) Biochem Biophys Res Commun 386: 159-164
- PubMed: 19505434
- DOI: https://doi.org/10.1016/j.bbrc.2009.06.004
- Primary Citation of Related Structures:
3HDB - PubMed Abstract:
AaHIV, a P-III-type snake venom metalloproteinase (SVMP), consists of metalloproteinase/disintegrin/cysteine-rich (MDC) domains and is homologous to a disintegrin and metalloproteinase (ADAM) family proteins. Similar to brevilysin H6 and jararhagin, AaHIV can easily autolyse to release a stable protein named acucetin, which contains disintegrin-like and cysteine-rich domains. In this study, we determined the crystal structure of AaHIV and investigated the autolysis mechanism. Based on the structure of AaHIV and the results from docking experiments, we present a new model for target recognition in which two protein molecules form a functional unit, and the DC domain of one molecule is used for target recognition while the M-domain of the other is used for target proteolysis. Our results shed new light on the mechanism of target recognition and processing in ADAM/reprolysin family proteins.
- Hefei National Laboratory for Physical Sciences at Microscale and School of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230026, China.
Organizational Affiliation: