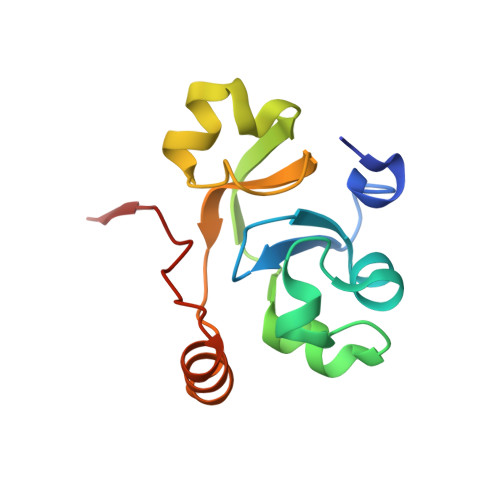
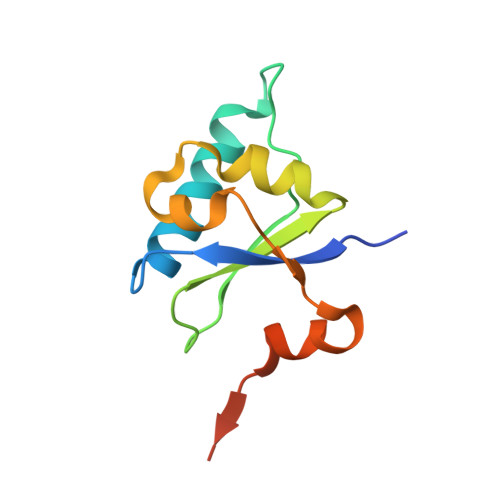
Crystal structure of the human transcription elongation factor DSIF hSpt4 subunit in complex with the hSpt5 dimerization interface
Wenzel, S., Martins, B.M., Rosch, P., Wohrl, B.M.(2010) Biochem J 425: 373-380
- PubMed: 19860741
- DOI: https://doi.org/10.1042/BJ20091422
- Primary Citation of Related Structures:
3H7H - PubMed Abstract:
The eukaryotic transcription elongation factor DSIF [DRB (5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole) sensitivity-inducing factor] is composed of two subunits, hSpt4 and hSpt5, which are homologous to the yeast factors Spt4 and Spt5. DSIF is involved in regulating the processivity of RNA polymerase II and plays an essential role in transcriptional activation of eukaryotes. At several eukaryotic promoters, DSIF, together with NELF (negative elongation factor), leads to promoter-proximal pausing of RNA polymerase II. In the present paper we describe the crystal structure of hSpt4 in complex with the dimerization region of hSpt5 (amino acids 176-273) at a resolution of 1.55 A (1 A=0.1 nm). The heterodimer shows high structural similarity to its homologue from Saccharomyces cerevisiae. Furthermore, hSpt5-NGN is structurally similar to the NTD (N-terminal domain) of the bacterial transcription factor NusG. A homologue for hSpt4 has not yet been found in bacteria. However, the archaeal transcription factor RpoE" appears to be distantly related. Although a comparison of the NusG-NTD of Escherichia coli with hSpt5 revealed a similarity of the three-dimensional structures, interaction of E. coli NusG-NTD with hSpt4 could not be observed by NMR titration experiments. A conserved glutamate residue, which was shown to be crucial for dimerization in yeast, is also involved in the human heterodimer, but is substituted for a glutamine residue in Escherichia coli NusG. However, exchanging the glutamine for glutamate proved not to be sufficient to induce hSpt4 binding.
- Research Centre for Biomacromolecules, Universitätsstr. 30, D-95447 Bayreuth, Germany.
Organizational Affiliation: