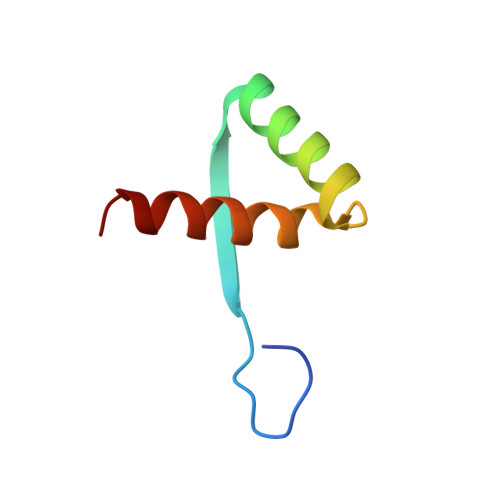
The Staphylococcus aureus pSK41 plasmid-encoded ArtA protein is a master regulator of plasmid transmission genes and contains a RHH motif used in alternate DNA-binding modes.
Ni, L., Jensen, S.O., Ky Tonthat, N., Berg, T., Kwong, S.M., Guan, F.H., Brown, M.H., Skurray, R.A., Firth, N., Schumacher, M.A.(2009) Nucleic Acids Res 37: 6970-6983
- PubMed: 19759211
- DOI: https://doi.org/10.1093/nar/gkp756
- Primary Citation of Related Structures:
3GXQ - PubMed Abstract:
Plasmids harbored by Staphylococcus aureus are a major contributor to the spread of bacterial multi-drug resistance. Plasmid conjugation and partition are critical to the dissemination and inheritance of such plasmids. Here, we demonstrate that the ArtA protein encoded by the S. aureus multi-resistance plasmid pSK41 is a global transcriptional regulator of pSK41 genes, including those involved in conjugation and segregation. ArtA shows no sequence homology to any structurally characterized DNA-binding protein. To elucidate the mechanism by which it specifically recognizes its DNA site, we obtained the structure of ArtA bound to its cognate operator, ACATGACATG. The structure reveals that ArtA is representative of a new family of ribbon-helix-helix (RHH) DNA-binding proteins that contain extended, N-terminal basic motifs. Strikingly, unlike most well-studied RHH proteins ArtA binds its cognate operators as a dimer. However, we demonstrate that it is also able to recognize an atypical operator site by binding as a dimer-of-dimers and the extended N-terminal regions of ArtA were shown to be essential for this dimer-of-dimer binding mode. Thus, these data indicate that ArtA is a master regulator of genes critical for both horizontal and vertical transmission of pSK41 and that it can recognize DNA utilizing alternate binding modes.
- Department of Biochemistry and Molecular Biology, University of Texas, MD Anderson Cancer Center, Unit 1000, Houston, TX 77030, USA.
Organizational Affiliation: