Structural diversity in calmodulin recognition of nitric oxide synthases
Ng, H.L., Greenstein, A., Marletta, M., Wand, A.J., Alber, T.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Calmodulin | 148 | Gallus gallus | Mutation(s): 0 Gene Names: CALM, CAM, RCJMB04_24e7 | 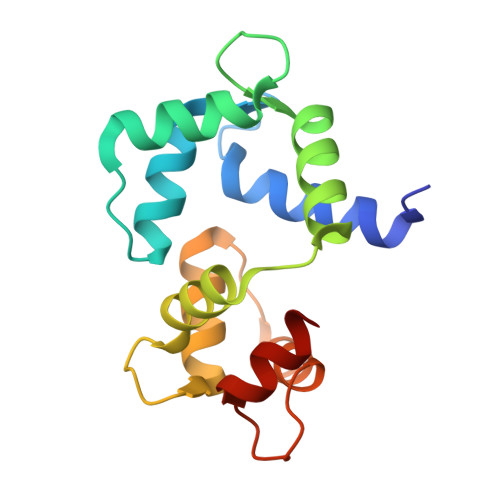 | |
UniProt | |||||
Find proteins for P62149 (Gallus gallus) Explore P62149 Go to UniProtKB: P62149 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62149 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Nitric oxide synthase, inducible | 16 | N/A | Mutation(s): 0 EC: 1.14.13.39 | 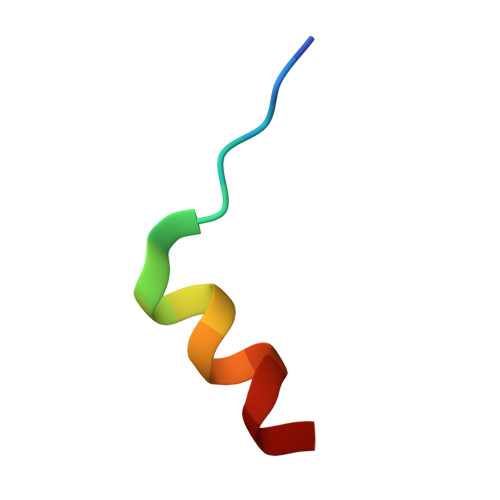 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P29477 (Mus musculus) Explore P29477 Go to UniProtKB: P29477 | |||||
IMPC: MGI:97361 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P29477 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SO4 Query on SO4 | I [auth A], N [auth B] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| CA Query on CA | E [auth A] F [auth A] G [auth A] H [auth A] J [auth B] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 44.923 | α = 90 |
| b = 57.773 | β = 93.06 |
| c = 60.231 | γ = 90 |
| Software Name | Purpose |
|---|---|
| Blu-Ice | data collection |
| SOLVE | phasing |
| REFMAC | refinement |
| MOSFLM | data reduction |
| SCALA | data scaling |