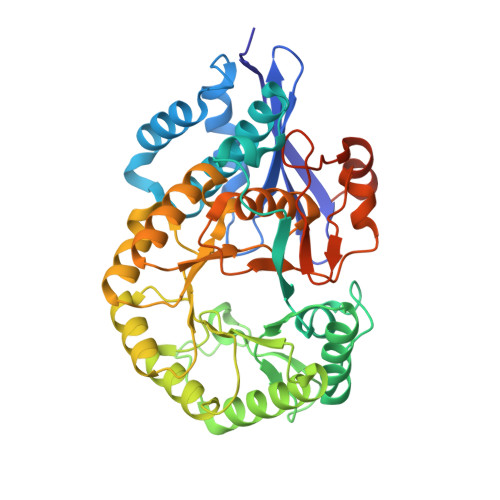
Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400 bound to magnesium.
Bonanno, J.B., Dickey, M., Bain, K.T., Chang, S., Ozyurt, S., Wasserman, S., Sauder, J.M., Burley, S.K., Almo, S.C.To be published.