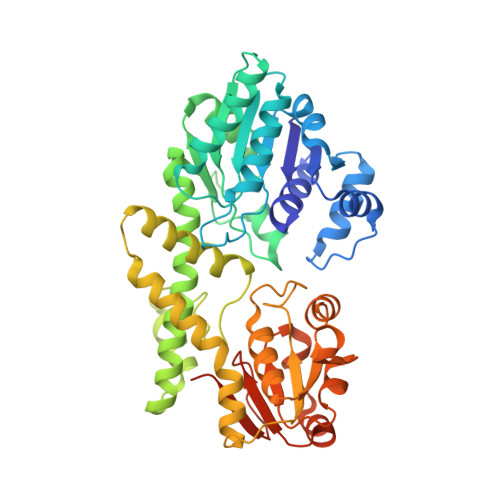
Crystal structure of UDP-glucose 6-dehydrogenase from Porphyromonas gingivalis bound to product UDP-glucuronate
Bonanno, J.B., Freeman, J., Bain, K.T., Chang, S., Sampathkumar, P., Wasserman, S., Sauder, J.M., Burley, S.K., Almo, S.C.To be published.