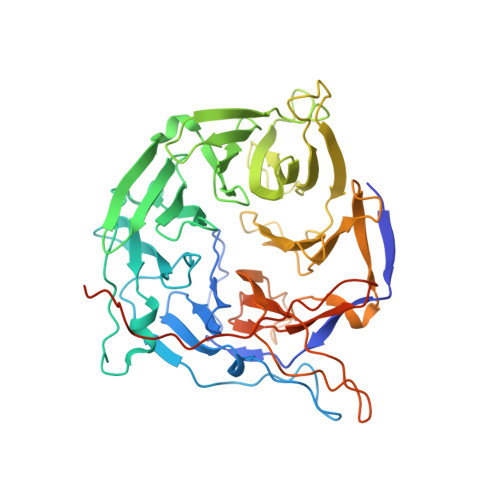
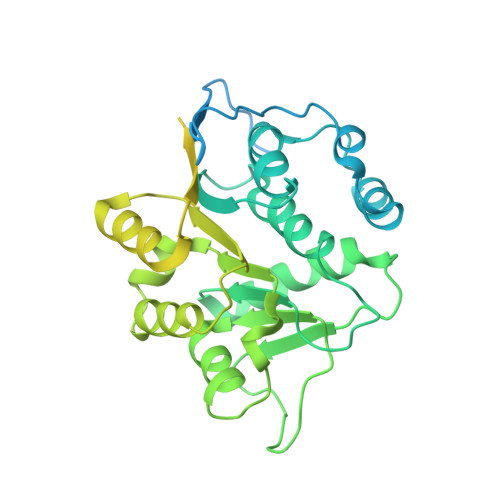
Structural and functional analysis of the interaction between the nucleoporin Nup214 and the DEAD-box helicase Ddx19.
Napetschnig, J., Kassube, S.A., Debler, E.W., Wong, R.W., Blobel, G., Hoelz, A.(2009) Proc Natl Acad Sci U S A 106: 3089-3094
- PubMed: 19208808
- DOI: https://doi.org/10.1073/pnas.0813267106
- Primary Citation of Related Structures:
3FMO, 3FMP - PubMed Abstract:
Key steps in the export of mRNA from the nucleus to the cytoplasm are the transport through the nuclear pore complex (NPC) and the subsequent remodeling of messenger RNA-protein (mRNP) complexes that occurs at the cytoplasmic side of the NPC. Crucial for these events is the recruitment of the DEAD-box helicase Ddx19 to the cytoplasmic filaments of the NPC that is mediated by the nucleoporin Nup214. Here, we present the crystal structure of the Nup214 N-terminal domain in complex with Ddx19 in its ADP-bound state at 2.5 A resolution. Strikingly, the interaction surfaces are not only evolutionarily conserved but also exhibit strongly opposing surface potentials, with the helicase surface being positively and the Nup214 surface being negatively charged. We speculate that the positively charged surface of the interacting ADP-helicase binds competitively to a segment of mRNA of a linearized mRNP, passing through the NPC on its way to the cytoplasm. As a result, the ADP-helicase would dissociate from Nup214 and replace a single bound protein from the mRNA. One cycle of protein replacement would be accompanied, cooperatively, by nucleotide exchange, ATP hydrolysis, release of the ADP-helicase from mRNA and its rebinding to Nup214. Repeat of these cycles would remove proteins from a mRNP, one at a time, akin to a ratchet mechanism for mRNA export.
- Laboratory of Cell Biology, Howard Hughes Medical Institute, The Rockefeller University, 1230 York Avenue, New York, NY 10065, USA.
Organizational Affiliation: