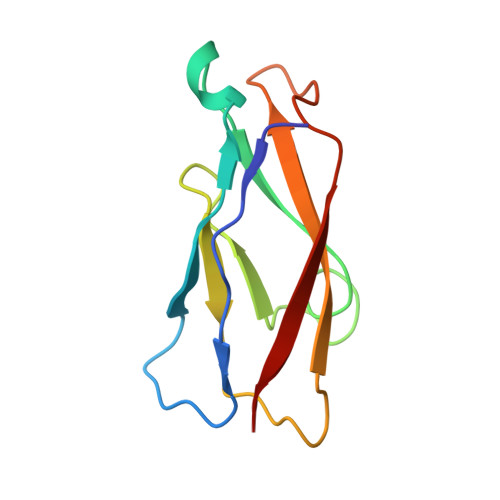
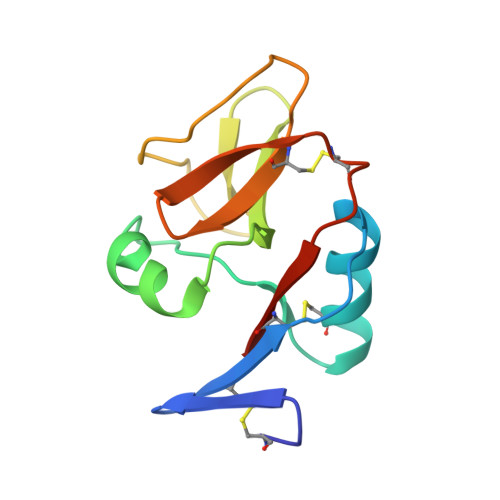
Structure of natural killer cell receptor KLRG1 bound to E-cadherin reveals basis for MHC-independent missing self recognition.
Li, Y., Hofmann, M., Wang, Q., Teng, L., Chlewicki, L.K., Pircher, H., Mariuzza, R.A.(2009) Immunity 31: 35-46
- PubMed: 19604491
- DOI: https://doi.org/10.1016/j.immuni.2009.04.019
- Primary Citation of Related Structures:
3FF7, 3FF8, 3FF9 - PubMed Abstract:
The cytolytic activity of natural killer (NK) cells is regulated by inhibitory receptors that detect the absence of self molecules on target cells. Structural studies of missing self recognition have focused on NK receptors that bind MHC. However, NK cells also possess inhibitory receptors specific for non-MHC ligands, notably cadherins, which are downregulated in metastatic tumors. We determined the structure of killer cell lectin-like receptor G1 (KLRG1) in complex with E-cadherin. KLRG1 mediates missing self recognition by binding to a highly conserved site on classical cadherins, enabling it to monitor expression of several cadherins (E-, N-, and R-) on target cells. This site overlaps the site responsible for cell-cell adhesion but is distinct from the integrin alpha(E)beta(7) binding site. We propose that E-cadherin may coengage KLRG1 and alpha(E)beta(7) and that KLRG1 overcomes its exceptionally weak affinity for cadherins through multipoint attachment to target cells, resulting in inhibitory signaling.
- Center for Advanced Research in Biotechnology, WM Keck Laboratory for Structural Biology, University of Maryland Biotechnology Institute, Rockville, MD 20850, USA.
Organizational Affiliation: