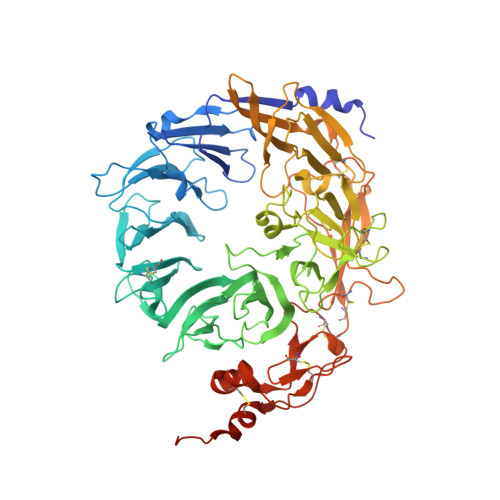
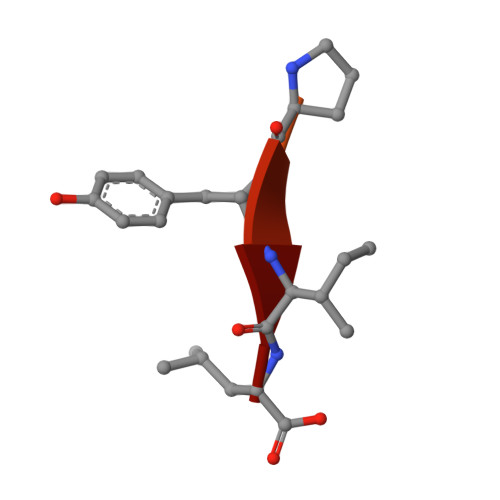
Ligands bind to Sortilin in the tunnel of a ten-bladed beta-propeller domain.
Quistgaard, E.M., Madsen, P., Groftehauge, M.K., Nissen, P., Petersen, C.M., Thirup, S.S.(2009) Nat Struct Mol Biol 16: 96-98
- PubMed: 19122660
- DOI: https://doi.org/10.1038/nsmb.1543
- Primary Citation of Related Structures:
3F6K - PubMed Abstract:
The structure of the Sortilin ectodomain in complex with neurotensin has been determined at 2-A resolution, revealing that the C-terminal part of neurotensin binds in the tunnel of a ten-bladed beta-propeller domain. Binding competition studies suggest that additional binding sites, for example, for the prodomain of nerve growth factor-beta, are present in the tunnel and that competition for binding relates to the restricted space inside the propeller.
- MIND Centre, Department of Molecular Biology, University of Aarhus, Gustav Wieds Vej 10C, DK 8000 Arhus C, Denmark.
Organizational Affiliation: