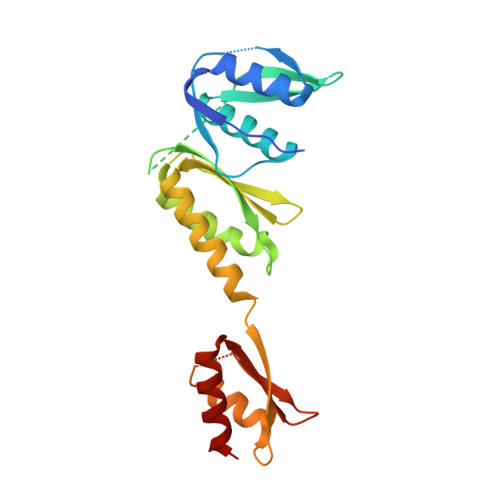
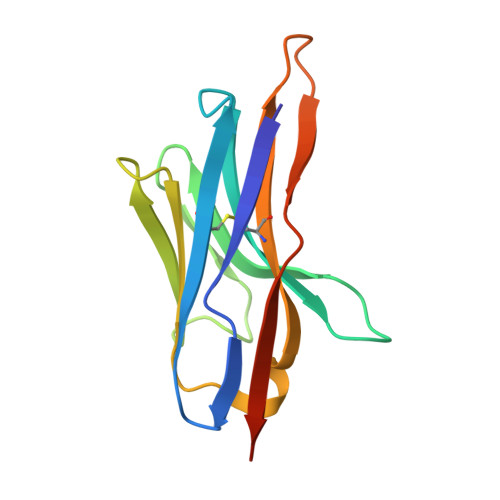
Crystal structure of the N-terminal domain of the secretin GspD from ETEC determined with the assistance of a nanobody.
Korotkov, K.V., Pardon, E., Steyaert, J., Hol, W.G.(2009) Structure 17: 255-265
- PubMed: 19217396
- DOI: https://doi.org/10.1016/j.str.2008.11.011
- Primary Citation of Related Structures:
3EZJ - PubMed Abstract:
Secretins are among the largest bacterial outer membrane proteins known. Here we report the crystal structure of the periplasmic N-terminal domain of GspD (peri-GspD) from the type 2 secretion system (T2SS) secretin in complex with a nanobody, the VHH domain of a heavy-chain camelid antibody. Two different crystal forms contained the same compact peri-GspD:nanobody heterotetramer. The nanobody contacts peri-GspD mainly via CDR3 and framework residues. The peri-GspD structure reveals three subdomains, with the second and third subdomains exhibiting the KH fold which also occurs in ring-forming proteins of the type 3 secretion system. The first subdomain of GspD is related to domains in phage tail proteins and outer membrane TonB-dependent receptors. A dodecameric peri-GspD model is proposed in which a solvent-accessible beta strand of the first subdomain interacts with secreted proteins and/or T2SS partner proteins by beta strand complementation.
- Department of Biochemistry, Biomolecular Structure Center, University of Washington, Seattle, WA 98195, USA.
Organizational Affiliation: