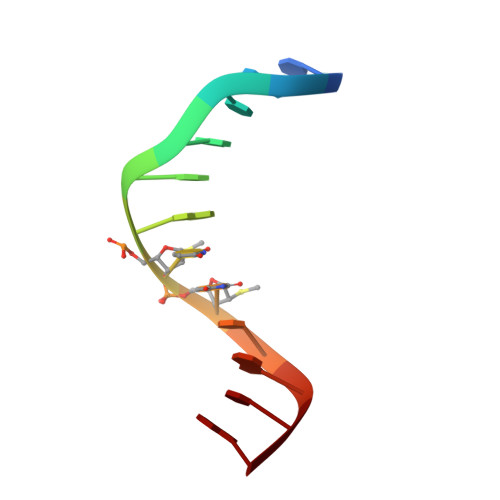
A conformational transition in the structure of a 2'-thiomethyl-modified DNA visualized at high resolution.
Pallan, P.S., Prakash, T.P., Li, F., Eoff, R.L., Manoharan, M., Egli, M.(2009) Chem Commun (Camb) 15: 2017-2019
- PubMed: 19333476
- DOI: https://doi.org/10.1039/b822781k
- Primary Citation of Related Structures:
3EY1, 3EY2, 3EY3 - PubMed Abstract:
Crystal structures of A-form and B-form DNA duplexes containing 2'-S-methyl-uridines reveal that the modified residues adopt a RNA-like C3'-endo pucker, illustrating that the replacement of electronegative oxygen at the 2'-carbon of RNA by sulfur does not appear to fundamentally alter the conformational preference of the sugar in the oligonucleotide context and sterics trump stereoelectronics.
- Department of Biochemistry, Vanderbilt University School of Medicine, Nashville, TN 37232, USA.
Organizational Affiliation: