Crystal structure of the full length eIF5A from Saccharomyces cerevisiae
Sanches, M., Dias, C.A.O., Aponi, L.H., Valentini, S.R., Guimaraes, B.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Eukaryotic translation initiation factor 5A-2 | 167 | Saccharomyces cerevisiae | Mutation(s): 0 Gene Names: HYP2, TIF51A, YEL034W, SYGP-ORF21 | 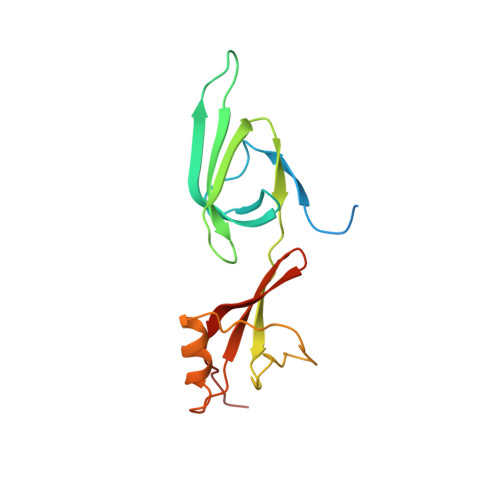 | |
UniProt | |||||
Find proteins for P23301 (Saccharomyces cerevisiae (strain ATCC 204508 / S288c)) Explore P23301 Go to UniProtKB: P23301 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P23301 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 59.664 | α = 90 |
| b = 59.664 | β = 90 |
| c = 339.79 | γ = 120 |
| Software Name | Purpose |
|---|---|
| MAR345dtb | data collection |
| PHASER | phasing |
| REFMAC | refinement |
| MOSFLM | data reduction |
| SCALA | data scaling |