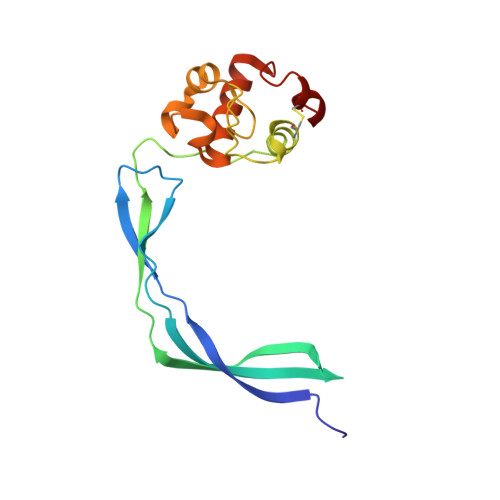
Crystal Structure of the Resuscitation-Promoting Factor (DeltaDUF)RpfB from M. tuberculosis
Ruggiero, A., Tizzano, B., Pedone, E., Pedone, C., Wilmanns, M., Berisio, R.(2009) J Mol Biology 385: 153-162
- PubMed: 18992255
- DOI: https://doi.org/10.1016/j.jmb.2008.10.042
- Primary Citation of Related Structures:
3EO5 - PubMed Abstract:
Mycobacterium tuberculosis is able to establish a non-replicating state and survive in an intracellular habitat for years. Resuscitation of dormant M. tuberculosis bacteria is promoted by resuscitation-promoting factors (Rpfs), which are secreted from slowly replicating bacteria close to dormant bacteria. Here we report the crystal structure of a truncated form of RpfB (residues 194-362), the sole indispensable Rpf of the five Rpfs encoded in this bacterium genome. The structure, denoted as (DeltaDUF)RpfB, exhibits a comma-like shape formed by a lysozyme-like globular catalytic domain and an elongated G5 domain, which is widespread among cell surface binding proteins. The G5 domain, whose structure was previously uncharacterised, presents some peculiar features. The basic structural motif of this domain, which represents the tail of the comma-like structure, is a novel super-secondary-structure element, made of two beta-sheets interconnected by a pseudo-triple helix. This intricate organisation leads to the exposure of several backbone hydrogen-bond donors/acceptors. Mutagenesis analyses and solution studies indicate that this protein construct as well as the full-length form are elongated monomeric proteins. Although (DeltaDUF)RpfB does not self-associate, the exposure of structural elements (backbone H-bond donors/acceptors and hydrophobic side chains) that are usually buried in globular proteins is typically associated with adhesive properties. This suggests that the RpfB G5 domain has a cell-wall adhesive function, which allows the catalytic domain to be properly oriented for the cleavage reaction. Interestingly, sequence comparisons indicate that these structural features are also shared by G5 domains involved in biofilm formation.
- Instituto di Biostrutture e Bioimmagini, CNR, Naples, Italy.
Organizational Affiliation: