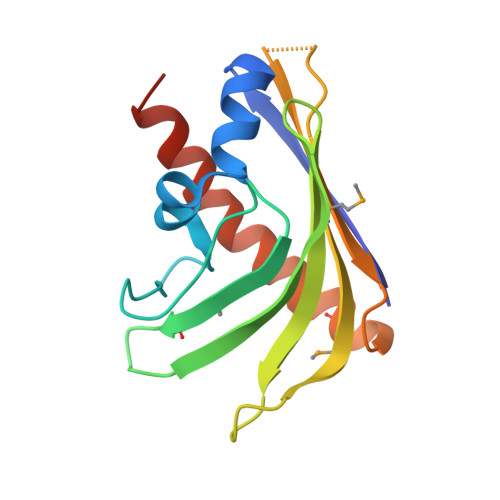
Crystal structure of the AHSA1 (SPO3351) protein from Silicibacter pomeroyi, Northeast Structural Genomics Consortium Target SiR160
Forouhar, F., Su, M., Seetharaman, J., Janjua, H., Xiao, R., Ciccosanti, C., Foote, E.L., Wang, D., Tong, S., Everett, J.K., Acton, T.B., Montelione, G.T., Hunt, J.F., Tong, L.To be published.