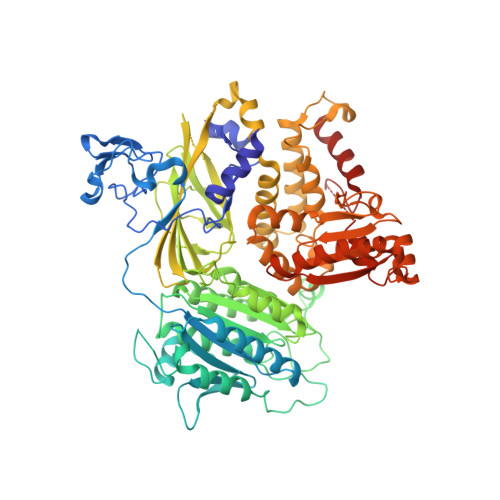
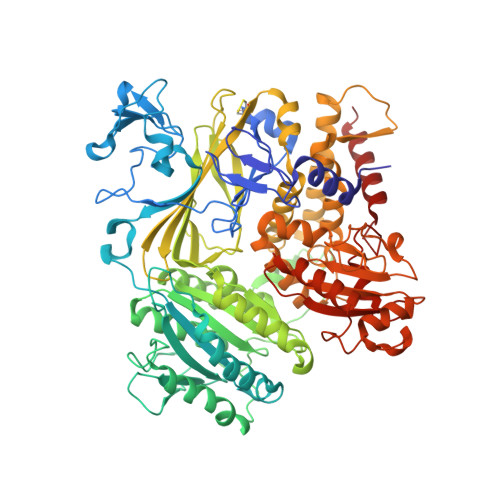
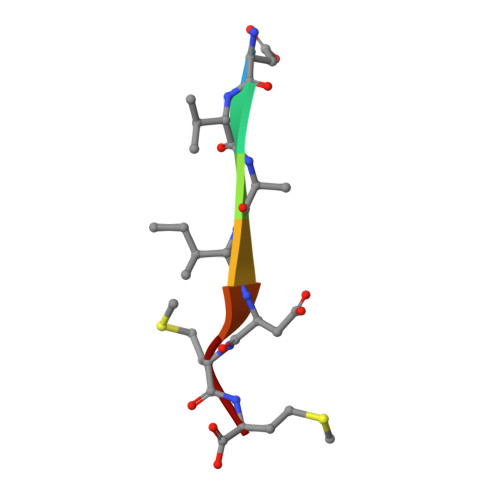
Structural basis of cargo membrane protein discrimination by the human COPII coat machinery.
Mancias, J.D., Goldberg, J.(2008) EMBO J 27: 2918-2928
- PubMed: 18843296
- DOI: https://doi.org/10.1038/emboj.2008.208
- Primary Citation of Related Structures:
3EFO, 3EG9, 3EGD, 3EGX, 3EH1, 3EH2 - PubMed Abstract:
Genomic analysis shows that the increased complexity of trafficking pathways in mammalian cells involves an expansion of the number of SNARE, Rab and COP proteins. Thus, the human genome encodes four forms of Sec24, the cargo selection subunit of the COPII vesicular coat, and this is proposed to increase the range of cargo accommodated by human COPII-coated vesicles. In this study, we combined X-ray crystallographic and biochemical analysis with functional assays of cargo packaging into COPII vesicles to establish molecular mechanisms for cargo discrimination by human Sec24 subunits. A conserved IxM packaging signal binds in a surface groove of Sec24c and Sec24d, but the groove is occluded in the Sec24a and Sec24b subunits. Conversely, LxxLE class transport signals and the DxE signal of VSV glycoprotein are selectively bound by Sec24a and Sec24b subunits. A comparative analysis of crystal structures of the four human Sec24 isoforms establishes the structural determinants for discrimination among these transport signals, and provides a framework to understand how an expansion of coat subunits extends the range of cargo proteins packaged into COPII-coated vesicles.
- Howard Hughes Medical Institute and the Structural Biology Program, Memorial Sloan-Kettering Cancer Center, New York, NY 10065, USA.
Organizational Affiliation: