Early Interrogation and Recognition of DNA Sequence by Indirect Readout
Little, E.J., Babic, A.C., Horton, N.C.(2008) Structure 16: 1828-1837
- PubMed: 19081059
- DOI: https://doi.org/10.1016/j.str.2008.09.009
- Primary Citation of Related Structures:
3EBC - PubMed Abstract:
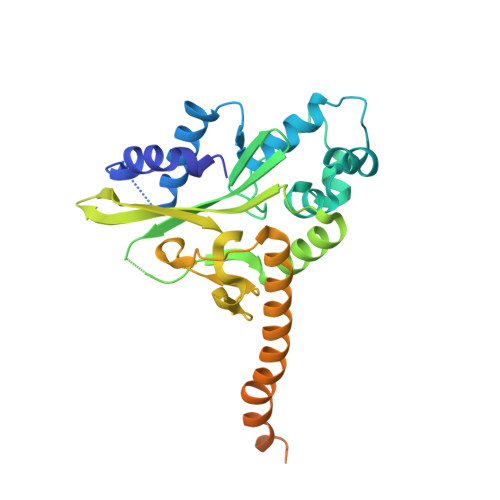
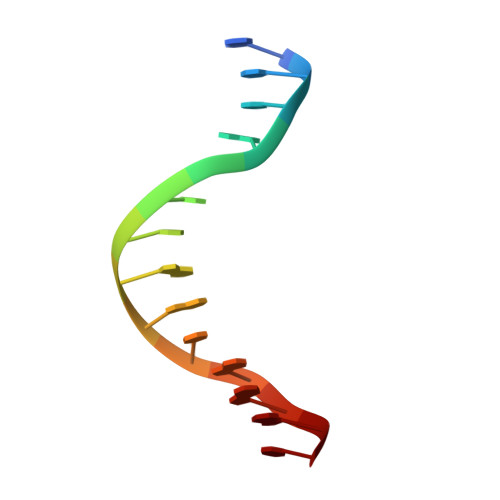
Control of replication, transcription, recombination and repair requires proteins capable of finding particular DNA sequences in a background of a large excess of nonspecific sequences. Such recognition can involve direct readout, with direct contacts to the bases of DNA, or in some cases through the less well-characterized indirect readout mechanisms. In order to measure the relative contributions of direct and indirect readout by a sequence specific endonuclease, HincII, a mutant enzyme deficient in a direct contact, was characterized, and surprisingly showed no loss of sequence specificity. The three dimensional crystal structure shows the loss of most of the direct readout contacts to the DNA, possibly capturing an early stage in target site recognition using predominately indirect readout to prescreen sites before full sequence interrogation.
- Department of Biochemistry and Molecular Biophysics, University of Arizona, Tucson, AZ 85721, USA.
Organizational Affiliation: