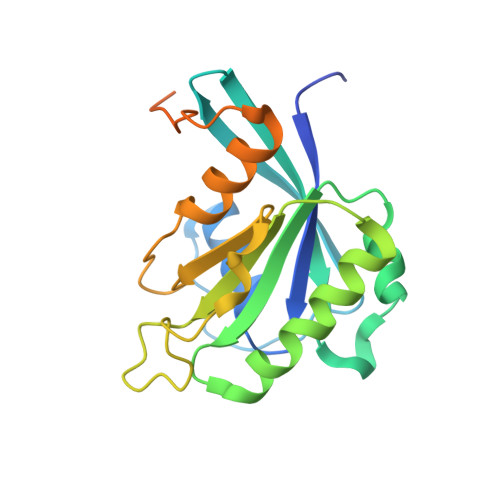
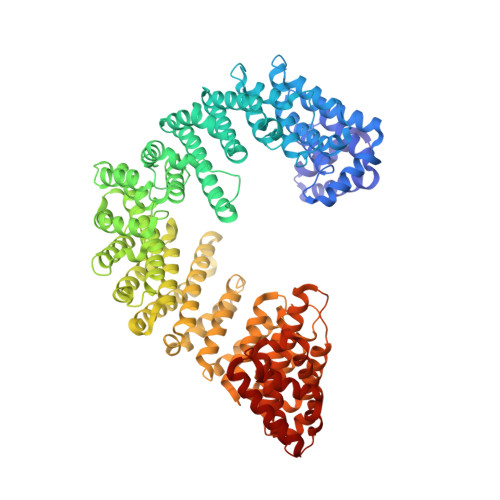
Kap95p binding induces the switch loops of RanGDP to adopt the GTP-bound conformation: implications for nuclear import complex assembly dynamics.
Forwood, J.K., Lonhienne, T.G., Marfori, M., Robin, G., Meng, W., Guncar, G., Liu, S.M., Stewart, M., Carroll, B.J., Kobe, B.(2008) J Mol Biology 383: 772-782
- PubMed: 18708071
- DOI: https://doi.org/10.1016/j.jmb.2008.07.090
- Primary Citation of Related Structures:
3EA5 - PubMed Abstract:
The asymmetric distribution of the nucleotide-bound state of Ran across the nuclear envelope is crucial for determining the directionality of nuclear transport. In the nucleus, Ran is primarily in the guanosine 5'-triphosphate (GTP)-bound state, whereas in the cytoplasm, Ran is primarily guanosine 5'-diphosphate (GDP)-bound. Conformational changes within the Ran switch I and switch II loops are thought to modulate its affinity for importin-beta. Here, we show that RanGDP and importin-beta form a stable complex with a micromolar dissociation constant. This complex can be dissociated by importin-beta binding partners such as importin-alpha. Surprisingly, the crystal structure of the Kap95p-RanGDP complex shows that Kap95p induces the switch I and II regions of RanGDP to adopt a conformation that resembles that of the GTP-bound form. The structure of the complex provides insights into the structural basis for the gradation of affinities regulating nuclear protein transport.
- School of Biomedical Sciences, Charles Sturt University, Wagga Wagga 2650, Australia. jforwood@csu.edu.au
Organizational Affiliation: