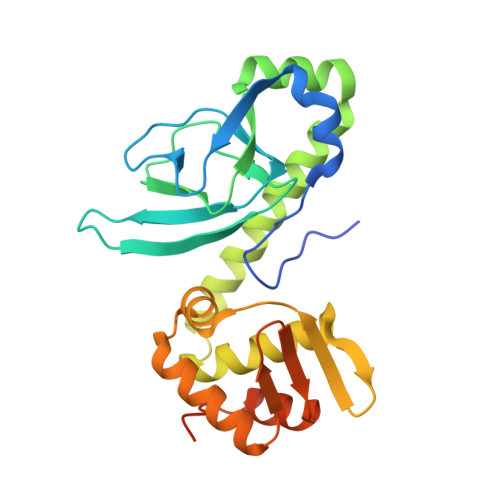
Molecular basis of halorespiration control by CprK, a CRP-FNR type transcriptional regulator
Levy, C., Pike, K., Heyes, D.J., Joyce, M.G., Gabor, K., Smidt, H., van der Oost, J., Leys, D.(2008) Mol Microbiol 70: 151-167
- PubMed: 18717788
- DOI: https://doi.org/10.1111/j.1365-2958.2008.06399.x
- Primary Citation of Related Structures:
3E5Q, 3E5U, 3E5X, 3E6B, 3E6C, 3E6D - PubMed Abstract:
Certain bacteria are able to conserve energy via the reductive dehalogenation of halo-organic compounds in a respiration-type metabolism. The transcriptional regulator CprK from Desulfitobacterium spp. induces expression of halorespiratory genes upon binding of o-chlorophenol ligands and is reversibly inactivated by oxygen through disulphide bond formation. We report crystal structures of D. hafniense CprK in the ligand-free (both oxidation states), ligand-bound (reduced) and DNA-bound states, making it the first member of the widespread CRP-FNR superfamily for which a complete structural description of both redox-dependent and allosteric molecular rearrangements is available. In conjunction with kinetic and thermodynamic ligand binding studies, we provide a model for the allosteric mechanisms underpinning transcriptional control. Amino acids that play a key role in this mechanism are not conserved in functionally distinct CRP-FNR members. This suggests that, despite significant structural homology, distinct allosteric mechanisms are used, enabling this protein family to control a very wide range of processes.
- Manchester Interdisciplinary Biocentre, The University of Manchester, Manchester M1 7DN, UK.
Organizational Affiliation: