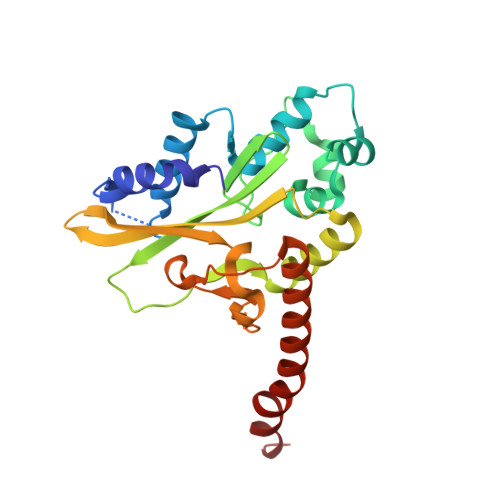
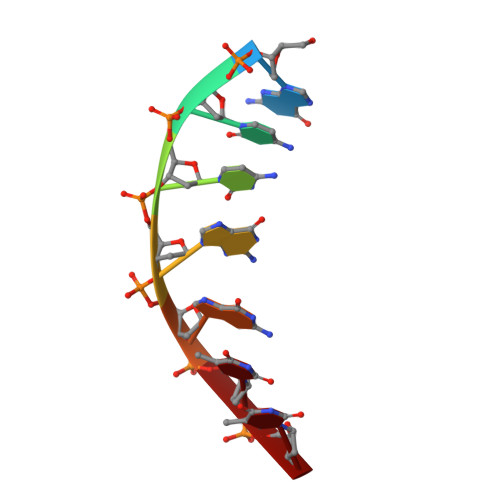
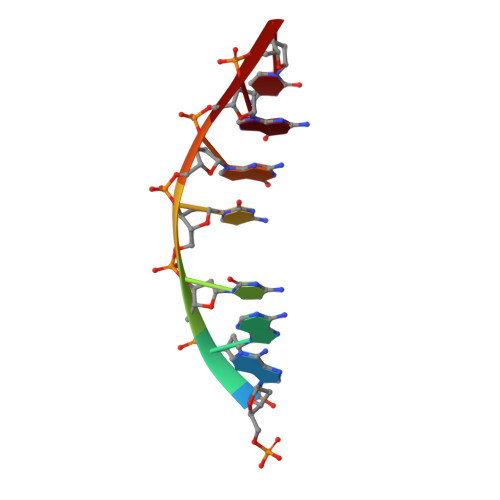
DNA distortion and specificity in a sequence-specific endonuclease.
Babic, A.C., Little, E.J., Manohar, V.M., Bitinaite, J., Horton, N.C.(2008) J Mol Biology 383: 186-204
- PubMed: 18762194
- DOI: https://doi.org/10.1016/j.jmb.2008.08.032
- Primary Citation of Related Structures:
3E3Y, 3E40, 3E41, 3E42, 3E43, 3E44, 3E45 - PubMed Abstract:
Five new structures of the Q138F HincII enzyme bound to a total of three different DNA sequences and three different metal ions (Ca(2+), Mg(2+), and Mn(2+)) are presented. While previous structures were produced from soaking Ca(2+) into preformed Q138F HincII/DNA crystals, the new structures are derived from cocrystallization with Ca(2+), Mg(2+), or Mn(2+). The Mn(2)(+)-bound structure provides the first view of a product complex of Q138F HincII with cleaved DNA. Binding studies and a crystal structure show how Ca(2+) allows trapping of a Q138F HincII complex with noncognate DNA in a catalytically incompetent conformation. Many Q138F HincII/DNA structures show asymmetry, despite the binding of a symmetric substrate by a symmetric enzyme. The various complexes are fit into a model describing the different conformations of the DNA-bound enzyme and show how DNA conformational energetics determine DNA-cleavage rates by the Q138F HincII enzyme.
- Department of Biochemistry and Molecular Biophysics, University of Arizona, Tucson, AZ 85721, USA.
Organizational Affiliation: