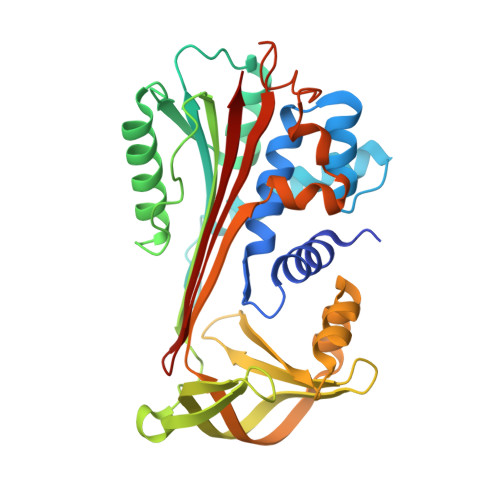
The heparin binding site of protein C inhibitor is protease-dependent.
Li, W., Huntington, J.A.(2008) J Biological Chem 283: 36039-36045
- PubMed: 18974053
- DOI: https://doi.org/10.1074/jbc.M805974200
- Primary Citation of Related Structures:
3DY0 - PubMed Abstract:
Protein C inhibitor (PCI) is a member of the serpin family of protease inhibitors with many biological functions and broad inhibitory specificity. Its major targets in blood are thrombin and activated protein C (APC), and the inhibition of both enzymes can be accelerated by glycosaminoglycans, including heparin. Acceleration of thrombin and APC inhibition by PCI requires that both protease and inhibitor bind to the same heparin chain to form a bridged Michaelis complex. However, the position of the heparin binding site of APC is opposite to that of thrombin, and formation of the bridged complexes must require either radical reorientation of the proteases relative to PCI or alternate heparin binding modes for PCI. In this study, we investigate how heparin bridges thrombin and APC to PCI by determining the effect of mutations in and around the putative heparin binding site of PCI. We found that heparin binds PCI in a linear fashion along helix H to bridge thrombin, consistent with our recent crystal structure (3B9F), but that it must rotate by approximately 60 degrees to engage Arg-229 to bridge APC. To gain insight into the possible modes of heparin binding to PCI, we solved a crystal structure of cleaved PCI bound to an octasaccharide heparin fragment to 1.55 angstroms resolution. The structure reveals a binding mode across the N terminus of helix H to engage Arg-229 and align the heparin binding site of APC. A molecular model for the heparin-bridged PCI.APC complex was built based on mutagenesis and structural data.
- Department of Haematology, University of Cambridge, Cambridge Institute for Medical Research, Wellcome Trust/MRC Building, Hills Road, Cambridge, CB2 0XY, United Kingdom.
Organizational Affiliation: