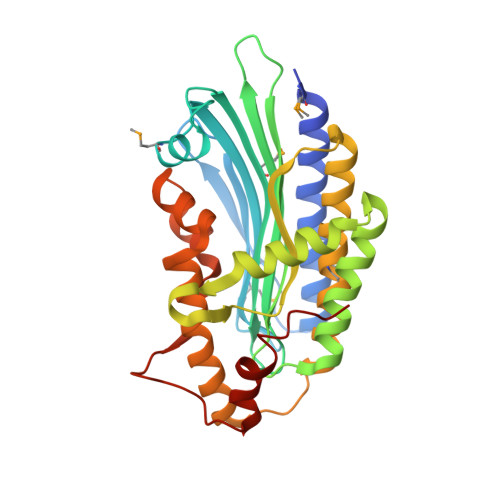
Leishmania major Coproporphyrinogen III Oxidase with bound ligand
merritt, E.A., Le Trong, I., Larson, E.T., Shibata, S., Zhang, Z., Anderson, L., Ross, J., Deng, W., Verlinde, C.L.M.J., Buckner, F.S., Van Voorhis, W.C., Hol, W.G.J., Fan, E.To be published.