Genetic and structural insights into the dissemination potential of the extremely broad-spectrum class A beta-lactamase KPC-2 identified in an Escherichia coli strain and an Enterobacter cloacae strain isolated from the same patient in France.
Petrella, S., Ziental-Gelus, N., Mayer, C., Renard, M., Jarlier, V., Sougakoff, W.(2008) Antimicrob Agents Chemother 52: 3725-3736
- PubMed: 18625772
- DOI: https://doi.org/10.1128/AAC.00163-08
- Primary Citation of Related Structures:
3DW0 - PubMed Abstract:
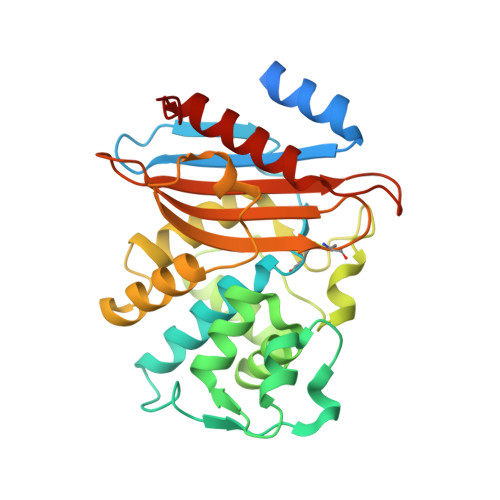
Two clinical strains of Escherichia coli (2138) and Enterobacter cloacae (7506) isolated from the same patient in France and showing resistance to extended-spectrum cephalosporins and low susceptibility to imipenem were investigated. Both strains harbored the plasmid-contained bla(TEM-1) and bla(KPC-2) genes. bla(KLUC-2), encoding a mutant of the chromosomal beta-lactamase of Kluyvera cryocrescens, was also identified at a plasmid location in E. cloacae 7506, suggesting the ISEcp1-assisted escape of bla(KLUC) from the chromosome. Determination of the KPC-2 structure at 1.6 A revealed that the binding site was occupied by the C-terminal (C-ter) residues coming from a symmetric KPC-2 monomer, with the ultimate C-ter Glu interacting with Ser130, Lys234, Thr235, and Thr237 in the active site. This mode of binding can be paralleled to the inhibition of the TEM-1 beta-lactamase by the inhibitory protein BLIP. Determination of the 1.23-A structure of a KPC-2 mutant in which the five C-ter residues were deleted revealed that the catalytic site was filled by a citrate molecule. Structure analysis and docking simulations with cefotaxime and imipenem provided further insights into the molecular basis of the extremely broad spectrum of KPC-2, which behaves as a cefotaximase with significant activity against carbapenems. In particular, residues 104, 105, 132, and 167 draw a binding cavity capable of accommodating both the aminothiazole moiety of cefotaxime and the 6 alpha-hydroxyethyl group of imipenem, with the binding of the former drug being also favored by a significant degree of freedom at the level of the loop at positions 96 to 105 and by an enlargement of the binding site at the end of strand beta 3.
- UPMC Univ Paris 06, EA1541, Bacteriologie-Hygiène, Paris, France.
Organizational Affiliation: