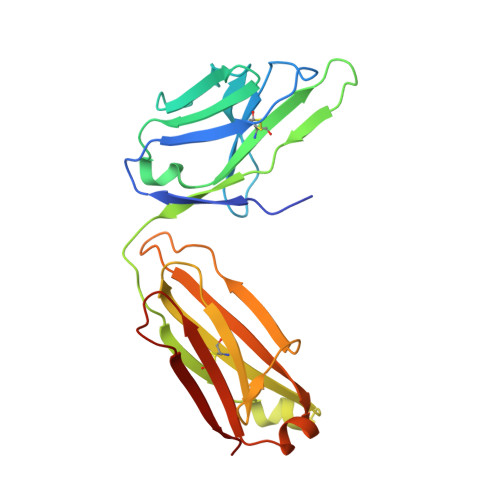
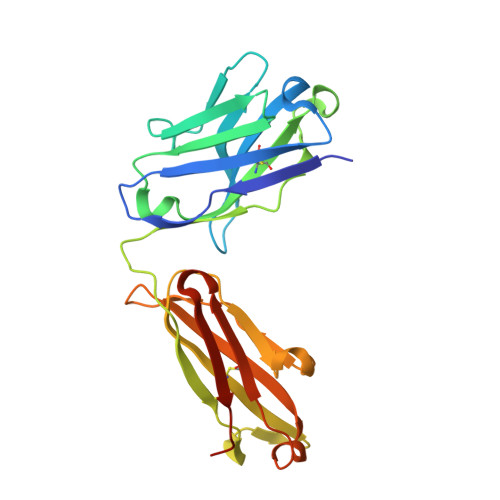
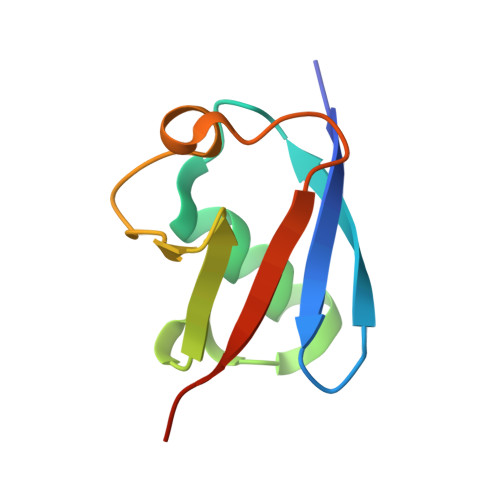
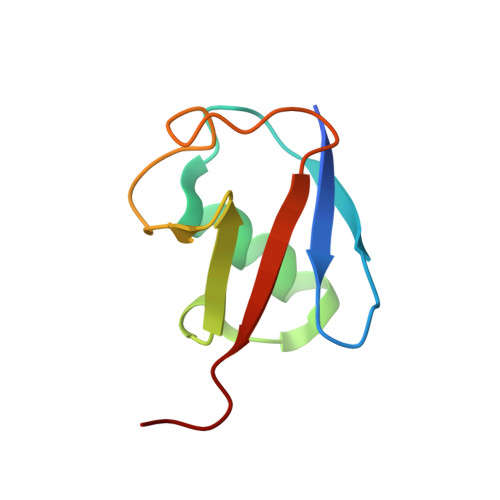
Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies.
Newton, K., Matsumoto, M.L., Wertz, I.E., Kirkpatrick, D.S., Lill, J.R., Tan, J., Dugger, D., Gordon, N., Sidhu, S.S., Fellouse, F.A., Komuves, L., French, D.M., Ferrando, R.E., Lam, C., Compaan, D., Yu, C., Bosanac, I., Hymowitz, S.G., Kelley, R.F., Dixit, V.M.(2008) Cell 134: 668-678
- PubMed: 18724939
- DOI: https://doi.org/10.1016/j.cell.2008.07.039
- Primary Citation of Related Structures:
3DVG, 3DVN - PubMed Abstract:
Posttranslational modification of proteins with polyubiquitin occurs in diverse signaling pathways and is tightly regulated to ensure cellular homeostasis. Studies employing ubiquitin mutants suggest that the fate of polyubiquitinated proteins is determined by which lysine within ubiquitin is linked to the C terminus of an adjacent ubiquitin. We have developed linkage-specific antibodies that recognize polyubiquitin chains joined through lysine 63 (K63) or 48 (K48). A cocrystal structure of an anti-K63 linkage Fab bound to K63-linked diubiquitin provides insight into the molecular basis for specificity. We use these antibodies to demonstrate that RIP1, which is essential for tumor necrosis factor-induced NF-kappaB activation, and IRAK1, which participates in signaling by interleukin-1beta and Toll-like receptors, both undergo polyubiquitin editing in stimulated cells. Both kinase adaptors initially acquire K63-linked polyubiquitin, while at later times K48-linked polyubiquitin targets them for proteasomal degradation. Polyubiquitin editing may therefore be a general mechanism for attenuating innate immune signaling.
- Department of Physiological Chemistry, Genentech, Inc., 1 DNA Way, South San Francisco, CA 94080, USA.
Organizational Affiliation: