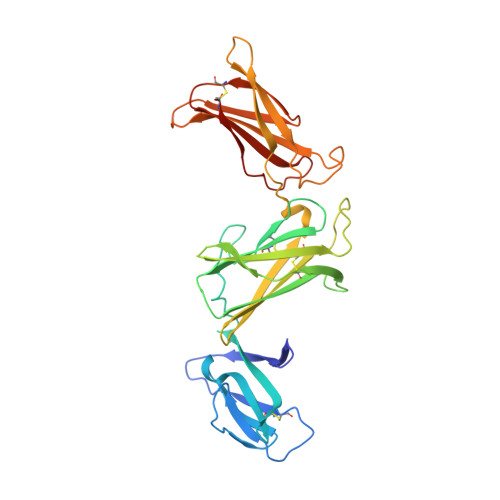
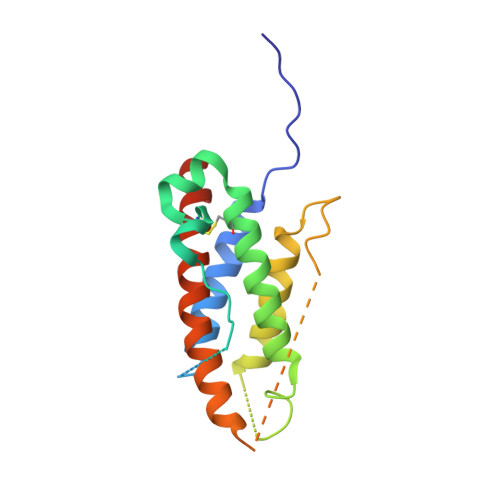
The structure of interleukin-23 reveals the molecular basis of p40 subunit sharing with interleukin-12.
Lupardus, P.J., Garcia, K.C.(2008) J Mol Biology 382: 931-941
- PubMed: 18680750
- DOI: https://doi.org/10.1016/j.jmb.2008.07.051
- Primary Citation of Related Structures:
3DUH - PubMed Abstract:
Interleukin (IL)-23 is a recently identified member of the IL-12 family of heterodimeric cytokines that modulate subpopulations of T helper cells, and both IL-12 and IL-23 are attractive targets for therapy of autoimmune diseases. IL-23 is a binary complex of a four-helix bundle cytokine (p19) and a soluble class I cytokine receptor p40. IL-12 and IL-23 share p40 as an alpha-receptor subunit, yet show only 15% sequence homology between their four-helix cytokines p19 and p35, respectively, and signal through different combinations of shared receptors. In order to elucidate the structural basis of p40 sharing, we have determined a 2.3-A crystal structure of IL-23 for comparison to the previously determined structure of IL-12. The docking mode of p19 to p40 is altered compared to p35, deviating by a 'tilt' and 'roll' that results in an altered footprint of p40 on the A and D helices of the respective cytokines. Binding of p19 to p40 is mediated primarily by an arginine residue on helix D of p19 that forms an extensive charge and hydrogen-bonding network with residues at the base of a pocket on p40. This 'arginine pocket' is lined with an inner shell of hydrophobic interactions that are ringed by an outer shell of polar interactions. Comparative analysis indicates that the IL-23 and IL-12 complexes 'mimic' the network of interactions constituting the central arginine pocket despite p19 and p35 having limited sequence homology. The majority of the structural epitopes in the two complexes are composed of unique p19 and p35 pairwise contacts with common residues on p40. Thus, while the critical hotspot is maintained in the two complexes, the majority of the interfaces are structurally distinct and, therefore, provide a basis for the therapeutic targeting of IL-12 versus IL-23 heterodimer formation despite their use of a common receptor subunit.
- Howard Hughes Medical Institute, Department of Molecular and Cellular Physiology, Stanford University School of Medicine, Stanford, CA 94305, USA.
Organizational Affiliation: