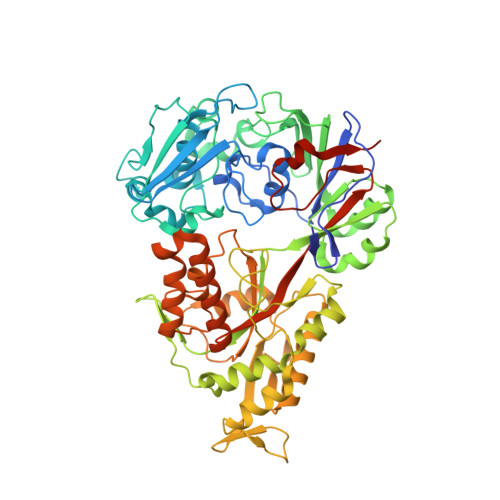
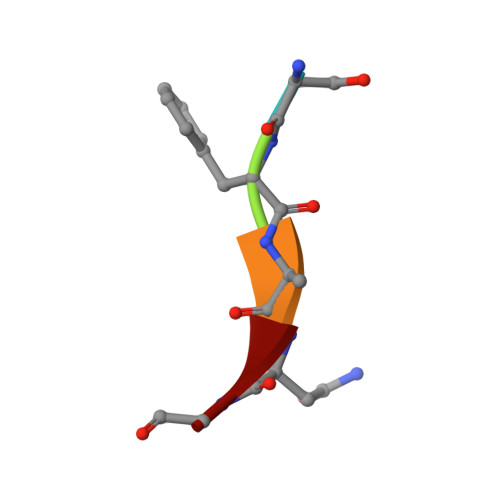
The structural basis for peptide selection by the transport receptor OppA
Berntsson, R.P.-A., Doeven, M.K., Fusetti, F., Duurkens, R.H., Sengupta, D., Marrink, S.-J., Thunnissen, A.-M., Poolman, B., Slotboom, D.-J.(2009) EMBO J 28: 1332-1340
- PubMed: 19300437
- DOI: https://doi.org/10.1038/emboj.2009.65
- Primary Citation of Related Structures:
3DRF, 3DRG, 3DRH, 3DRI, 3DRJ, 3DRK - PubMed Abstract:
Oligopeptide-binding protein A (OppA) from Lactococcus lactis binds peptides of an exceptionally wide range of lengths (4-35 residues), with no apparent sequence preference. Here, we present the crystal structures of OppA in the open- and closed-liganded conformations. The structures directly explain the protein's phenomenal promiscuity. A huge cavity allows binding of very long peptides, and a lack of constraints for the position of the N and C termini of the ligand is compatible with binding of peptides with varying lengths. Unexpectedly, the peptide's amino-acid composition (but not the exact sequence) appears to have a function in selection, with a preference for proline-rich peptides containing at least one isoleucine. These properties can be related to the physiology of the organism: L. lactis is auxotrophic for branched chain amino acids and favours proline-rich caseins as a source of amino acids. We propose a new mechanism for peptide selection based on amino-acid composition rather than sequence.
- Biochemistry Department, Groningen Biomolecular Sciences and Biotechnology Institute & Zernike Institute for Advanced Materials, University of Groningen, Groningen, The Netherlands.
Organizational Affiliation: